25 Vehicles classiification with Decision Trees
- Datasets: Vehicle (mlbench)
- Algorithms:
- Decision Trees
- Instructions: book “Applied Predictive Modeling Techniques”, Lewis, N.D.
25.1 Load packages
library(tree)
library(mlbench)
data(Vehicle)
str(Vehicle)
#> 'data.frame': 846 obs. of 19 variables:
#> $ Comp : num 95 91 104 93 85 107 97 90 86 93 ...
#> $ Circ : num 48 41 50 41 44 57 43 43 34 44 ...
#> $ D.Circ : num 83 84 106 82 70 106 73 66 62 98 ...
#> $ Rad.Ra : num 178 141 209 159 205 172 173 157 140 197 ...
#> $ Pr.Axis.Ra : num 72 57 66 63 103 50 65 65 61 62 ...
#> $ Max.L.Ra : num 10 9 10 9 52 6 6 9 7 11 ...
#> $ Scat.Ra : num 162 149 207 144 149 255 153 137 122 183 ...
#> $ Elong : num 42 45 32 46 45 26 42 48 54 36 ...
#> $ Pr.Axis.Rect: num 20 19 23 19 19 28 19 18 17 22 ...
#> $ Max.L.Rect : num 159 143 158 143 144 169 143 146 127 146 ...
#> $ Sc.Var.Maxis: num 176 170 223 160 241 280 176 162 141 202 ...
#> $ Sc.Var.maxis: num 379 330 635 309 325 957 361 281 223 505 ...
#> $ Ra.Gyr : num 184 158 220 127 188 264 172 164 112 152 ...
#> $ Skew.Maxis : num 70 72 73 63 127 85 66 67 64 64 ...
#> $ Skew.maxis : num 6 9 14 6 9 5 13 3 2 4 ...
#> $ Kurt.maxis : num 16 14 9 10 11 9 1 3 14 14 ...
#> $ Kurt.Maxis : num 187 189 188 199 180 181 200 193 200 195 ...
#> $ Holl.Ra : num 197 199 196 207 183 183 204 202 208 204 ...
#> $ Class : Factor w/ 4 levels "bus","opel","saab",..: 4 4 3 4 1 1 1 4 4 3 ...
summary(Vehicle[1])
#> Comp
#> Min. : 73.0
#> 1st Qu.: 87.0
#> Median : 93.0
#> Mean : 93.7
#> 3rd Qu.:100.0
#> Max. :119.0
summary(Vehicle[2])
#> Circ
#> Min. :33.0
#> 1st Qu.:40.0
#> Median :44.0
#> Mean :44.9
#> 3rd Qu.:49.0
#> Max. :59.0
attributes(Vehicle$Class)
#> $levels
#> [1] "bus" "opel" "saab" "van"
#>
#> $class
#> [1] "factor"25.3 Estimate the decision tree
fit <- tree(Class ~., data = trainset, split = "deviance")
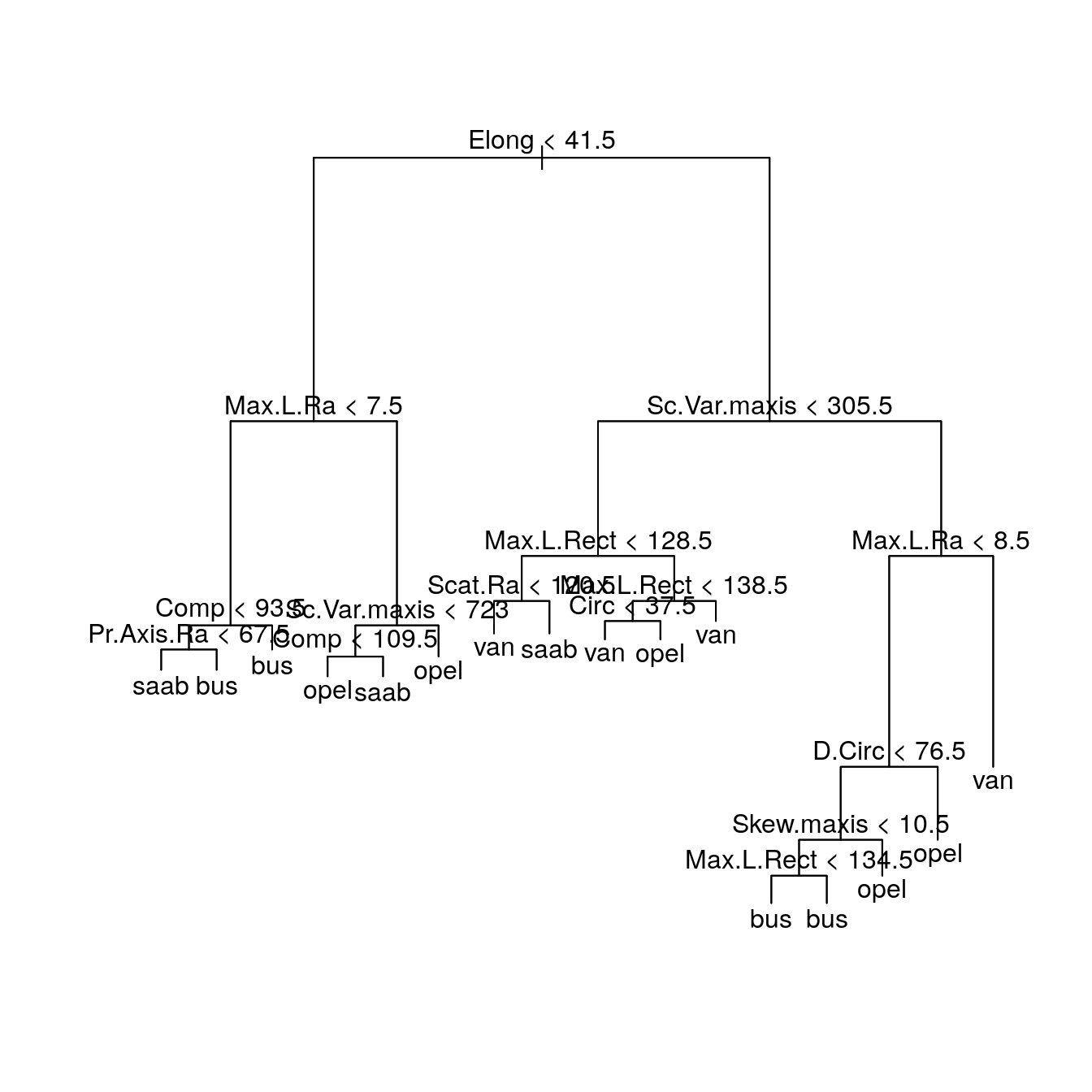
fit
#> node), split, n, deviance, yval, (yprob)
#> * denotes terminal node
#>
#> 1) root 500 1000 opel ( 0 0 0 0 )
#> 2) Elong < 41.5 215 500 saab ( 0 0 0 0 )
#> 4) Max.L.Ra < 7.5 51 50 bus ( 1 0 0 0 )
#> 8) Comp < 93.5 12 20 bus ( 0 0 0 0 )
#> 16) Pr.Axis.Ra < 67.5 7 8 saab ( 0 0 1 0 ) *
#> 17) Pr.Axis.Ra > 67.5 5 0 bus ( 1 0 0 0 ) *
#> 9) Comp > 93.5 39 9 bus ( 1 0 0 0 ) *
#> 5) Max.L.Ra > 7.5 164 200 opel ( 0 1 0 0 )
#> 10) Sc.Var.maxis < 723 149 200 saab ( 0 0 1 0 )
#> 20) Comp < 109.5 137 200 opel ( 0 1 0 0 ) *
#> 21) Comp > 109.5 12 0 saab ( 0 0 1 0 ) *
#> 11) Sc.Var.maxis > 723 15 7 opel ( 0 1 0 0 ) *
#> 3) Elong > 41.5 285 700 van ( 0 0 0 0 )
#> 6) Sc.Var.maxis < 305.5 116 200 van ( 0 0 0 1 )
#> 12) Max.L.Rect < 128.5 40 90 saab ( 0 0 0 0 )
#> 24) Scat.Ra < 120.5 15 30 van ( 0 0 0 1 ) *
#> 25) Scat.Ra > 120.5 25 30 saab ( 0 0 1 0 ) *
#> 13) Max.L.Rect > 128.5 76 90 van ( 0 0 0 1 )
#> 26) Max.L.Rect < 138.5 38 60 van ( 0 0 0 1 )
#> 52) Circ < 37.5 17 10 van ( 0 0 0 1 ) *
#> 53) Circ > 37.5 21 40 opel ( 0 0 0 0 ) *
#> 27) Max.L.Rect > 138.5 38 20 van ( 0 0 0 1 ) *
#> 7) Sc.Var.maxis > 305.5 169 400 bus ( 0 0 0 0 )
#> 14) Max.L.Ra < 8.5 116 200 bus ( 1 0 0 0 )
#> 28) D.Circ < 76.5 97 100 bus ( 1 0 0 0 )
#> 56) Skew.maxis < 10.5 87 70 bus ( 1 0 0 0 )
#> 112) Max.L.Rect < 134.5 12 20 bus ( 0 0 0 0 ) *
#> 113) Max.L.Rect > 134.5 75 20 bus ( 1 0 0 0 ) *
#> 57) Skew.maxis > 10.5 10 20 opel ( 0 0 0 0 ) *
#> 29) D.Circ > 76.5 19 30 opel ( 0 1 0 0 ) *
#> 15) Max.L.Ra > 8.5 53 20 van ( 0 0 0 1 ) *
# fit <- tree(Class ~., data = Vehicle[train,], split ="deviance")
# fitWe use deviance as the splitting criteria, a common alternative is to use split=“gini”.
At each branch of the tree (after root) we see in order: 1. The branch number (e.g. in this case 1,2,14 and 15); 2. the split (e.g. Elong < 41.5); 3. the number of samples going along that split (e.g. 229); 4. the deviance associated with that split (e.g. 489.1); 5. the predicted class (e.g. opel); 6. the associated probabilities (e.g. ( 0.222707 0.410480 0.366812 0.000000 ); 7. and for a terminal node (or leaf), the symbol "*".
summary(fit)
#>
#> Classification tree:
#> tree(formula = Class ~ ., data = trainset, split = "deviance")
#> Variables actually used in tree construction:
#> [1] "Elong" "Max.L.Ra" "Comp" "Pr.Axis.Ra" "Sc.Var.maxis"
#> [6] "Max.L.Rect" "Scat.Ra" "Circ" "D.Circ" "Skew.maxis"
#> Number of terminal nodes: 16
#> Residual mean deviance: 0.943 = 456 / 484
#> Misclassification error rate: 0.252 = 126 / 500Notice that summary(fit) shows: 1. The type of tree, in this case a Classification tree; 2. the formula used to fit the tree; 3. the variables used to fit the tree; 4. the number of terminal nodes in this case 15; 5. the residual mean deviance - 0.9381; 6. the misclassification error rate 0.232 or 23.2%.
25.4 Assess model
Unfortunately, classification trees have a tendency to overfit the data. One
approach to reduce this risk is to use cross-validation. For each hold out
sample we fit the model and note at what level the tree gives the best results
(using deviance or the misclassification rate). Then we hold out a different
sample and repeat. This can be carried out using the cv.tree() function.
We use a leave-one-out cross-validation using the misclassification rate and
deviance (FUN=prune.misclass, followed by FUN=prune.tree).
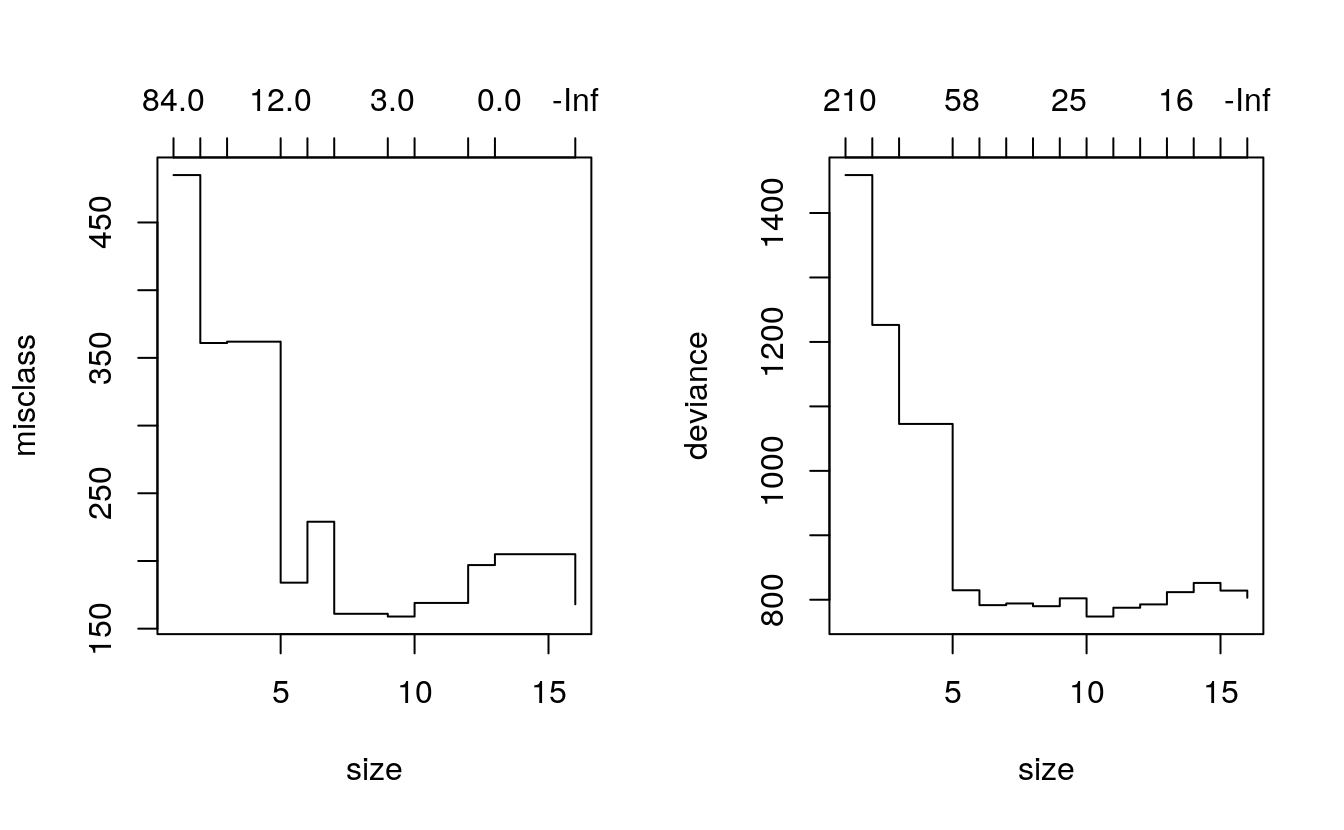
fitM.cv <- cv.tree(fit, K=346, FUN = prune.misclass)
fitP.cv <- cv.tree(fit, K=346, FUN = prune.tree)The results are plotted out side by side in Figure 1.2. The jagged lines shows where the minimum deviance / misclassification occurred with the cross-validated tree. Since the cross validated misclassification and deviance both reach their minimum close to the number of branches in the original fitted tree there is little to be gained from pruning this tree
25.5 Make predictions
We use the validation data set and the fitted decision tree to predict vehicle classes; then we display the confusion matrix and calculate the error rate of the fitted tree. Overall, the model has an error rate of 32%.
testLabels <- Vehicle$Class[-train]
testLabels
#> [1] van bus bus van van bus bus saab opel bus van saab van saab saab
#> [16] van saab opel van saab saab saab bus bus saab opel bus opel bus opel
#> [31] van opel opel saab saab bus bus bus van van saab opel bus opel van
#> [46] opel saab bus van bus opel van saab bus opel bus opel opel van bus
#> [61] van saab opel bus van saab opel opel saab saab saab opel bus van bus
#> [76] opel bus saab bus bus bus opel opel van saab bus bus bus van saab
#> [91] opel van van bus bus opel bus opel saab opel bus opel bus saab van
#> [106] van saab saab bus van opel van saab opel saab saab van van van van
#> [121] bus bus opel bus bus van saab bus opel bus bus bus bus opel van
#> [136] saab saab bus opel van bus saab bus van bus opel van saab opel saab
#> [151] opel van saab van saab opel bus van bus saab saab opel opel bus bus
#> [166] opel van van bus van van saab bus saab opel saab opel bus bus bus
#> [181] saab bus opel opel saab saab saab van van opel opel van van opel bus
#> [196] saab bus van opel opel bus bus bus opel saab opel van bus opel opel
#> [211] saab opel bus opel opel opel van opel van saab saab van saab saab saab
#> [226] saab van van van saab bus van van bus saab opel saab saab opel saab
#> [241] saab saab saab van saab opel bus saab bus opel opel opel saab bus van
#> [256] opel saab opel bus bus saab van opel bus saab van opel saab saab saab
#> [271] saab van opel bus bus bus opel saab saab saab van saab bus opel saab
#> [286] van opel bus saab saab opel opel van saab bus opel bus van van opel
#> [301] bus bus saab bus van saab bus van saab van opel bus bus opel saab
#> [316] opel bus bus saab van saab saab bus opel opel opel bus saab bus van
#> [331] bus van saab opel saab van opel opel van bus saab saab van saab opel
#> [346] saab
#> Levels: bus opel saab van
# Confusion Matrix
pred <- predict(fit, newdata = testset)
# find column whih has the maximum of all rows
pred.class <- colnames(pred)[max.col(pred, ties.method = c("random"))]
cm <- table(testLabels, pred.class,
dnn = c("Observed Class", "Predicted Class"))
cm
#> Predicted Class
#> Observed Class bus opel saab van
#> bus 85 1 1 5
#> opel 3 70 10 2
#> saab 7 67 14 7
#> van 1 4 5 64
# pred <- predict(fit, newdata = Vehicle[-train,])
# pred.class <- colnames(pred)[max.col(pred, ties.method = c("random"))]
# table(Vehicle$Class[-train], pred.class,
# dnn = c("Observed Class", "Predicted Class"))
# error_rate = (1 - sum(pred.class == Vehicle$Class[-train])/346)
# round(error_rate,3)