8 Ten methods to assess Variable Importance
Datasets:
GlaucomaM-
Algorithms:
- Partition Trees
- Regularized Random Forest (RRF)
- Lasso Regression
- Linear Regression
- Recursive Feature Elimination (RFE)
- Genetic Algorithm
- Simulated Annealing
Source: https://www.machinelearningplus.com/machine-learning/feature-selection/
In real-world datasets, it is fairly common to have columns that are nothing but noise.
You are better off getting rid of such variables because of the memory space they occupy, the time and the computational esources it is going to cost, especially in large datasets.
Sometimes, you have a variable that makes business sense, but you are not sure if it actually helps in predicting the Y. You also need to consider the fact that, a feature that could be useful in one ML algorithm (say a decision tree) may go underrepresented or unused by another (like a regression model).
Having said that, it is still possible that a variable that shows poor signs of helping to explain the response variable (Y), can turn out to be significantly useful in the presence of (or combination with) other predictors. What I mean by that is, a variable might have a low correlation value of (~0.2) with Y. But in the presence of other variables, it can help to explain certain patterns/phenomenon that other variables can’t explain.
In such cases, it can be hard to make a call whether to include or exclude such variables.
The strategies we are about to discuss can help fix such problems. Not only that, it will also help understand if a particular variable is important or not and how much it is contributing to the model
An important caveat. It is always best to have variables that have sound business logic backing the inclusion of a variable and rely solely on variable importance metrics.
Alright. Let’s load up the ‘Glaucoma’ dataset where the goal is to predict if a patient has Glaucoma or not based on 63 different physiological measurements. You can directly run the codes or download the dataset here.
A lot of interesting examples ahead. Let’s get started.
# Load Packages and prepare dataset
library(DALEX)
#> Welcome to DALEX (version: 1.2.0).
#> Find examples and detailed introduction at: https://pbiecek.github.io/ema/
library(TH.data)
#> Loading required package: survival
#> Loading required package: MASS
#>
#> Attaching package: 'TH.data'
#> The following object is masked from 'package:MASS':
#>
#> geyser
library(caret)
#> Loading required package: lattice
#> Loading required package: ggplot2
#>
#> Attaching package: 'caret'
#> The following object is masked from 'package:survival':
#>
#> cluster
library(tictoc)
data("GlaucomaM", package = "TH.data")
trainData <- GlaucomaM
head(trainData)
#> ag at as an ai eag eat eas ean eai abrg abrt abrs
#> 2 2.22 0.354 0.580 0.686 0.601 1.267 0.336 0.346 0.255 0.331 0.479 0.260 0.107
#> 43 2.68 0.475 0.672 0.868 0.667 2.053 0.440 0.520 0.639 0.454 1.090 0.377 0.257
#> 25 1.98 0.343 0.508 0.624 0.504 1.200 0.299 0.396 0.259 0.246 0.465 0.209 0.112
#> 65 1.75 0.269 0.476 0.525 0.476 0.612 0.147 0.017 0.044 0.405 0.170 0.062 0.000
#> 70 2.99 0.599 0.686 1.039 0.667 2.513 0.543 0.607 0.871 0.492 1.800 0.431 0.494
#> 16 2.92 0.483 0.763 0.901 0.770 2.200 0.462 0.637 0.504 0.597 1.311 0.394 0.365
#> abrn abri hic mhcg mhct mhcs mhcn mhci phcg phct phcs
#> 2 0.014 0.098 0.214 0.111 0.412 0.036 0.105 -0.022 -0.139 0.242 -0.053
#> 43 0.212 0.245 0.382 0.140 0.338 0.104 0.080 0.109 -0.015 0.296 -0.015
#> 25 0.041 0.103 0.195 0.062 0.356 0.045 -0.009 -0.048 -0.149 0.206 -0.092
#> 65 0.000 0.108 -0.030 -0.015 0.074 -0.084 -0.050 0.035 -0.182 -0.097 -0.125
#> 70 0.601 0.274 0.383 0.089 0.233 0.145 0.023 0.007 -0.131 0.163 0.055
#> 16 0.251 0.301 0.442 0.128 0.375 0.049 0.111 0.052 -0.088 0.281 -0.067
#> phcn phci hvc vbsg vbst vbss vbsn vbsi vasg vast vass vasn
#> 2 0.010 -0.139 0.613 0.303 0.103 0.088 0.022 0.090 0.062 0.000 0.011 0.032
#> 43 -0.015 0.036 0.382 0.676 0.181 0.186 0.141 0.169 0.029 0.001 0.007 0.011
#> 25 -0.081 -0.149 0.557 0.300 0.084 0.088 0.046 0.082 0.036 0.002 0.004 0.016
#> 65 -0.138 -0.182 0.373 0.048 0.011 0.000 0.000 0.036 0.070 0.005 0.030 0.033
#> 70 -0.131 -0.115 0.405 0.889 0.151 0.253 0.330 0.155 0.020 0.001 0.004 0.008
#> 16 -0.062 -0.088 0.507 0.972 0.213 0.316 0.197 0.246 0.043 0.001 0.005 0.028
#> vasi vbrg vbrt vbrs vbrn vbri varg vart vars varn vari mdg
#> 2 0.018 0.075 0.039 0.021 0.002 0.014 0.756 0.009 0.209 0.298 0.240 0.705
#> 43 0.010 0.370 0.127 0.099 0.050 0.093 0.410 0.006 0.105 0.181 0.117 0.898
#> 25 0.013 0.081 0.034 0.019 0.007 0.021 0.565 0.014 0.132 0.243 0.177 0.687
#> 65 0.002 0.005 0.001 0.000 0.000 0.004 0.380 0.032 0.147 0.151 0.050 0.207
#> 70 0.007 0.532 0.103 0.173 0.181 0.075 0.228 0.011 0.026 0.105 0.087 0.721
#> 16 0.009 0.467 0.136 0.148 0.078 0.104 0.540 0.008 0.133 0.232 0.167 0.927
#> mdt mds mdn mdi tmg tmt tms tmn tmi mr rnf mdic
#> 2 0.637 0.738 0.596 0.691 -0.236 -0.018 -0.230 -0.510 -0.158 0.841 0.410 0.137
#> 43 0.850 0.907 0.771 0.940 -0.211 -0.014 -0.165 -0.317 -0.192 0.924 0.256 0.252
#> 25 0.643 0.689 0.684 0.700 -0.185 -0.097 -0.235 -0.337 -0.020 0.795 0.378 0.152
#> 65 0.171 0.022 0.046 0.221 -0.148 -0.035 -0.449 -0.217 -0.091 0.746 0.200 0.027
#> 70 0.638 0.730 0.730 0.640 -0.052 -0.105 0.084 -0.012 -0.054 0.977 0.193 0.297
#> 16 0.842 0.953 0.906 0.898 -0.040 0.087 0.018 -0.094 -0.051 0.965 0.339 0.333
#> emd mv Class
#> 2 0.239 0.035 normal
#> 43 0.329 0.022 normal
#> 25 0.250 0.029 normal
#> 65 0.078 0.023 normal
#> 70 0.354 0.034 normal
#> 16 0.442 0.028 normal8.1 1. Boruta
Boruta is a feature ranking and selection algorithm based on random
forests algorithm. The advantage with Boruta is that it clearly decides
if a variable is important or not and helps to select variables that are
statistically significant. Besides, you can adjust the strictness of the
algorithm by adjusting the \(p\) values that defaults to 0.01 and the
maxRuns.
maxRuns is the number of times the algorithm is run. The higher the
maxRuns the more selective you get in picking the variables. The
default value is 100.
In the process of deciding if a feature is important or not, some features may be marked by Boruta as ‘Tentative’. Sometimes increasing the maxRuns can help resolve the ‘Tentativeness’ of the feature.
Lets see an example based on the Glaucoma dataset from TH.data package
that I created earlier.
The boruta function uses a formula interface just like most predictive
modeling functions. So the first argument to boruta() is the formula
with the response variable on the left and all the predictors on the
right. By placing a dot, all the variables in trainData other than
Class will be included in the model.
The doTrace argument controls the amount of output printed to the
console. Higher the value, more the log details you get. So save space I
have set it to 0, but try setting it to 1 and 2 if you are running the
code.
Finally the output is stored in boruta_output.
Let’s see what the boruta_output contains.
names(boruta_output)
#> [1] "finalDecision" "ImpHistory" "pValue" "maxRuns"
#> [5] "light" "mcAdj" "timeTaken" "roughfixed"
#> [9] "call" "impSource"
# Get significant variables including tentatives
boruta_signif <- getSelectedAttributes(boruta_output, withTentative = TRUE)
print(boruta_signif)
#> [1] "as" "ai" "eas" "ean" "abrg" "abrs" "abrn" "abri" "hic" "mhcg"
#> [11] "mhcs" "mhcn" "mhci" "phcg" "phcn" "phci" "hvc" "vbsg" "vbss" "vbsn"
#> [21] "vbsi" "vasg" "vass" "vasi" "vbrg" "vbrs" "vbrn" "vbri" "varg" "vart"
#> [31] "vars" "varn" "vari" "mdn" "tmg" "tmt" "tms" "tmi" "mr" "rnf"
#> [41] "mdic" "emd"If you are not sure about the tentative variables being selected for
granted, you can choose a TentativeRoughFix on boruta_output.
# Do a tentative rough fix
roughFixMod <- TentativeRoughFix(boruta_output)
boruta_signif <- getSelectedAttributes(roughFixMod)
print(boruta_signif)
#> [1] "as" "ai" "ean" "abrg" "abrs" "abrn" "abri" "hic" "mhcg" "mhcn"
#> [11] "mhci" "phcg" "phcn" "phci" "hvc" "vbsn" "vbsi" "vasg" "vass" "vasi"
#> [21] "vbrg" "vbrs" "vbrn" "vbri" "varg" "vart" "vars" "varn" "vari" "mdn"
#> [31] "tmg" "tms" "tmi" "mr" "rnf" "mdic"There you go. Boruta has decided on the ‘Tentative’ variables on our behalf. Let’s find out the importance scores of these variables.
# Variable Importance Scores
imps <- attStats(roughFixMod)
imps2 = imps[imps$decision != 'Rejected', c('meanImp', 'decision')]
head(imps2[order(-imps2$meanImp), ]) # descending sort
#> meanImp decision
#> vari 12.37 Confirmed
#> varg 11.74 Confirmed
#> vars 10.74 Confirmed
#> phci 8.34 Confirmed
#> hic 8.21 Confirmed
#> varn 7.88 ConfirmedLet’s plot it to see the importances of these variables.
# Plot variable importance
plot(boruta_output, cex.axis=.7, las=2, xlab="", main="Variable Importance") 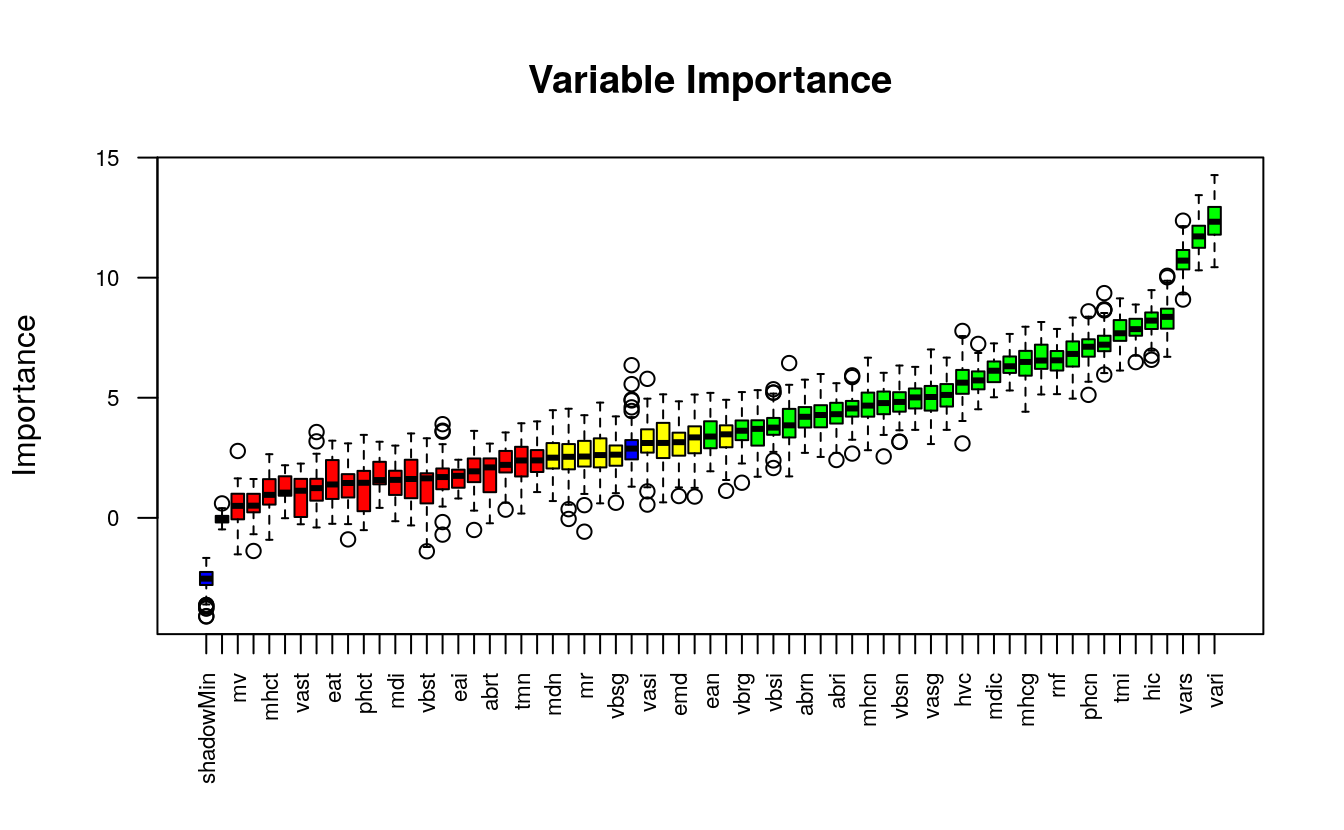
This plot reveals the importance of each of the features.
The columns in green are ‘confirmed’ and the ones in red are not. There
are couple of blue bars representing ShadowMax and ShadowMin. They
are not actual features, but are used by the boruta algorithm to
decide if a variable is important or not.
8.2 2. Variable Importance
Another way to look at feature selection is to consider variables most used by various ML algorithms the most to be important.
Depending on how the machine learning algorithm learns the relationship between \(X\)’s and $Y$, different machine learning algorithms may possibly end up using different variables (but mostly common vars) to various degrees.
What I mean by that is, the variables that proved useful in a tree-based
algorithm like rpart, can turn out to be less useful in a
regression-based model. So all variables need not be equally useful to
all algorithms.
So how do we find the variable importance for a given ML algo?
train() the desired model using the caret package. Then, use
varImp() to determine the feature importance.
You may want to try out multiple algorithms, to get a feel of the usefulness of the features across algos.
8.3 3. rpart
# Train an rpart model and compute variable importance.
library(caret)
set.seed(100)
rPartMod <- train(Class ~ .,
data=trainData,
method="rpart")
rpartImp <- varImp(rPartMod)
print(rpartImp)
#> rpart variable importance
#>
#> only 20 most important variables shown (out of 62)
#>
#> Overall
#> varg 100.0
#> vari 93.2
#> vars 85.2
#> varn 76.9
#> tmi 72.3
#> mhcn 0.0
#> as 0.0
#> phcs 0.0
#> vbst 0.0
#> abrt 0.0
#> vbsg 0.0
#> eai 0.0
#> vbrs 0.0
#> vbsi 0.0
#> eag 0.0
#> tmt 0.0
#> phcn 0.0
#> vart 0.0
#> mds 0.0
#> an 0.0Only 5 of the 63 features was used by rpart and if you look closely, the 5 variables used here are in the top 6 that boruta selected.
Let’s do one more: the variable importances from Regularized Random Forest (RRF) algorithm.
8.4 4. Regularized Random Forest (RRF)
tic()
# Train an RRF model and compute variable importance.
set.seed(100)
rrfMod <- train(Class ~ .,
data = trainData,
method = "RRF")
#> Registered S3 method overwritten by 'RRF':
#> method from
#> plot.margin randomForest
rrfImp <- varImp(rrfMod, scale=F)
toc()
#> 372.797 sec elapsed
rrfImp
#> RRF variable importance
#>
#> only 20 most important variables shown (out of 62)
#>
#> Overall
#> varg 25.07
#> vari 18.78
#> vars 5.29
#> tmi 4.09
#> mhcg 3.25
#> mhci 2.81
#> hic 2.69
#> hvc 2.50
#> mv 2.00
#> vasg 1.99
#> phci 1.77
#> phcn 1.53
#> phct 1.43
#> vass 1.37
#> phcg 1.37
#> tms 1.32
#> tmg 1.16
#> abrs 1.16
#> tmt 1.13
#> mdic 1.13
plot(rrfImp, top = 20, main='Variable Importance')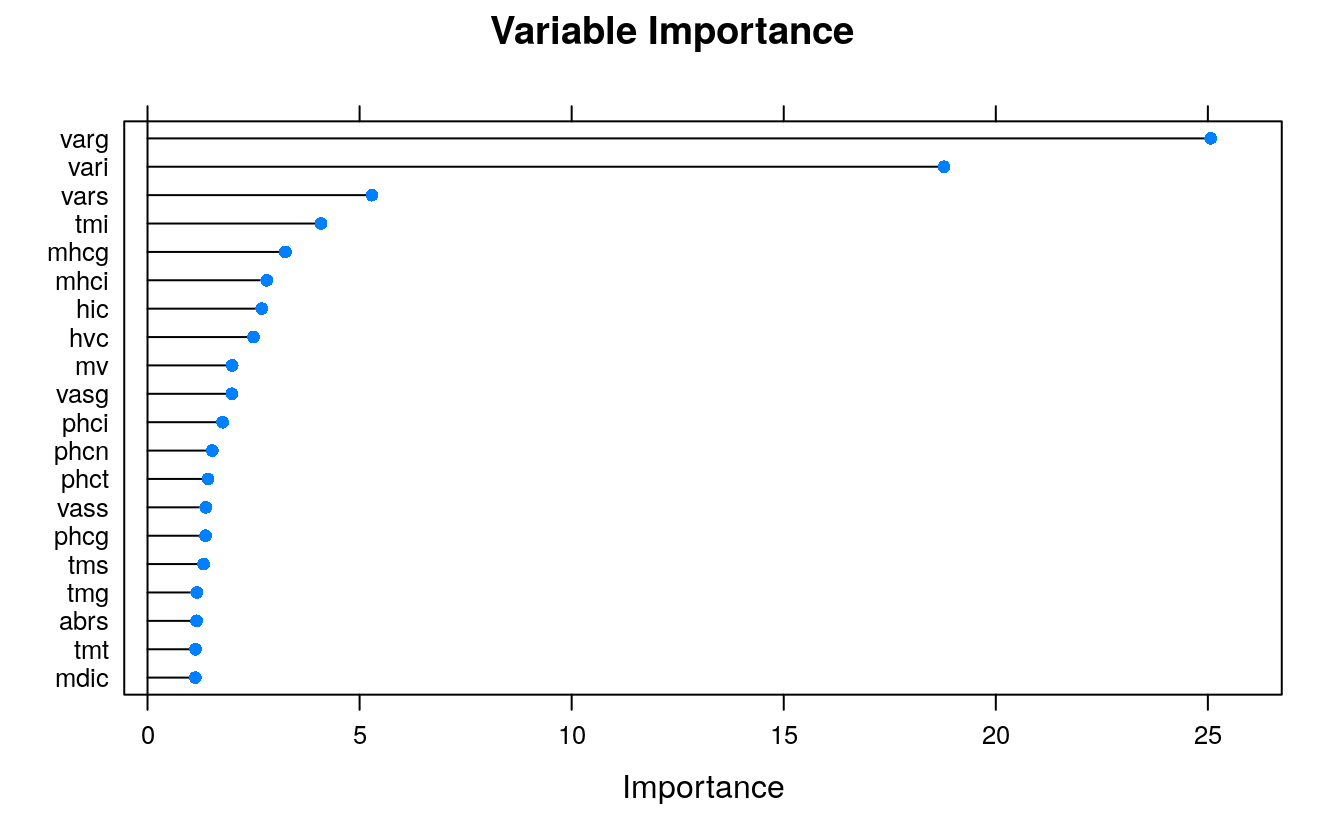
The topmost important variables are pretty much from the top tier of Boruta’s selections.
Some of the other algorithms available in train() that you can use to
compute varImp are the following:
ada, AdaBag, AdaBoost.M1, adaboost, bagEarth, bagEarthGCV, bagFDA, bagFDAGCV, bartMachine, blasso, BstLm, bstSm, C5.0, C5.0Cost, C5.0Rules, C5.0Tree, cforest, chaid, ctree, ctree2, cubist, deepboost, earth, enet, evtree, extraTrees, fda, gamboost, gbm_h2o, gbm, gcvEarth, glmnet_h2o, glmnet, glmStepAIC, J48, JRip, lars, lars2, lasso, LMT, LogitBoost, M5, M5Rules, msaenet, nodeHarvest, OneR, ordinalNet, ORFlog, ORFpls, ORFridge, ORFsvm, pam, parRF, PART, penalized, PenalizedLDA, qrf, ranger, Rborist, relaxo, rf, rFerns, rfRules, rotationForest, rotationForestCp, rpart, rpart1SE, rpart2, rpartCost, rpartScore, rqlasso, rqnc, RRF, RRFglobal, sdwd, smda, sparseLDA, spikeslab, wsrf, xgbLinear, xgbTree.8.5 5. Lasso Regression
Least Absolute Shrinkage and Selection Operator (LASSO) regression is a type of regularization method that penalizes with L1-norm.
It basically imposes a cost to having large weights (value of coefficients). And its called L1 regularization, because the cost added, is proportional to the absolute value of weight coefficients.
As a result, in the process of shrinking the coefficients, it eventually reduces the coefficients of certain unwanted features all the to zero. That is, it removes the unneeded variables altogether.
So effectively, LASSO regression can be considered as a variable selection technique as well.
library(glmnet)
#> Loading required package: Matrix
#> Loaded glmnet 3.0-2
# online data
# trainData <- read.csv('https://raw.githubusercontent.com/selva86/datasets/master/GlaucomaM.csv')
trainData <- read.csv(file.path(data_raw_dir, "glaucoma.csv"))
x <- as.matrix(trainData[,-63]) # all X vars
y <- as.double(as.matrix(ifelse(trainData[, 63]=='normal', 0, 1))) # Only Class
# Fit the LASSO model (Lasso: Alpha = 1)
set.seed(100)
cv.lasso <- cv.glmnet(x, y, family='binomial', alpha=1, parallel=TRUE, standardize=TRUE, type.measure='auc')
#> Warning: executing %dopar% sequentially: no parallel backend registered
# Results
plot(cv.lasso)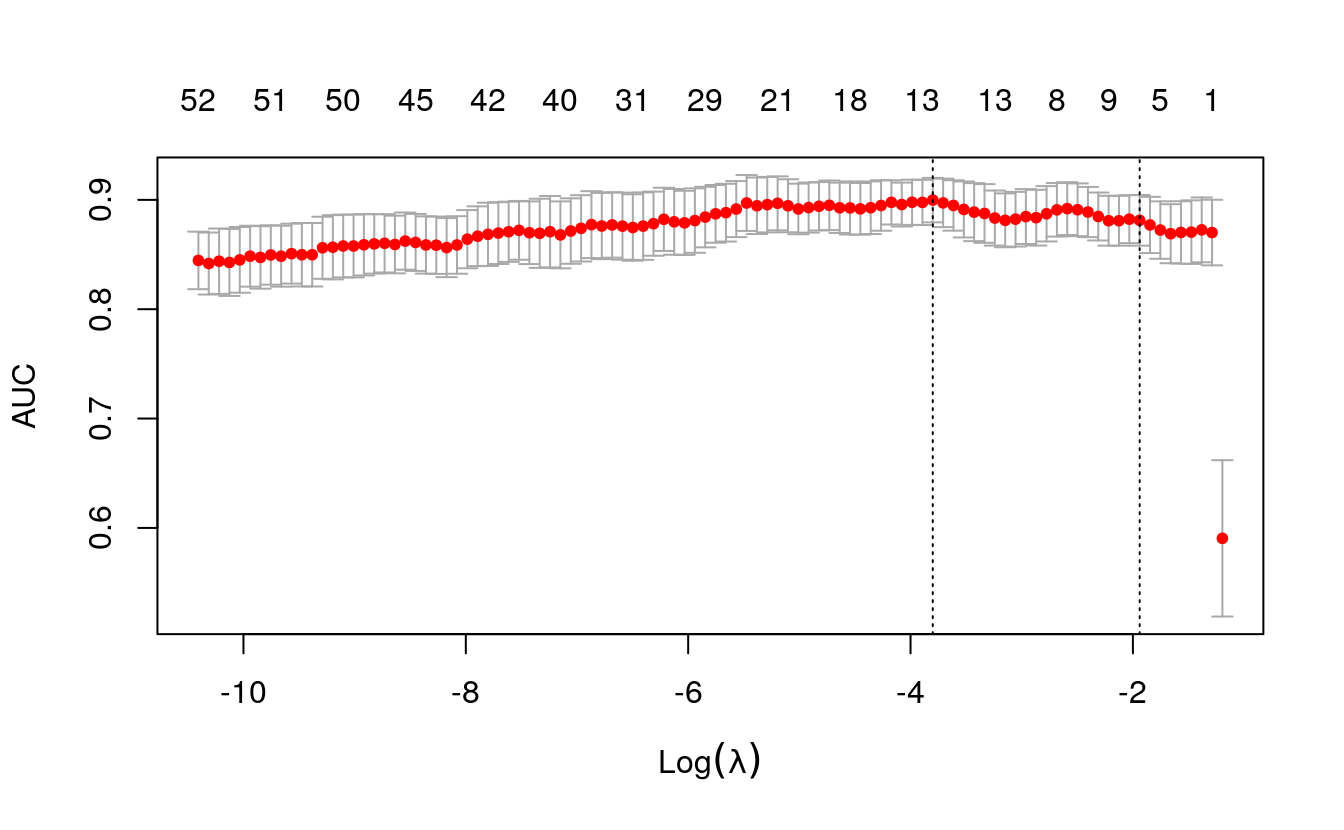
Let’s see how to interpret this plot.
The X axis of the plot is the log of lambda. That means when it is 2
here, the lambda value is actually 100.
The numbers at the top of the plot show how many predictors were
included in the model. The position of red dots along the Y-axis tells
what AUC we got when you include as many variables shown on the top
x-axis.
You can also see two dashed vertical lines.
The first one on the left points to the lambda with the lowest mean squared error. The one on the right point to the number of variables with the highest deviance within 1 standard deviation.
The best lambda value is stored inside ‘cv.lasso$lambda.min’.
# plot(cv.lasso$glmnet.fit, xvar="lambda", label=TRUE)
cat('Min Lambda: ', cv.lasso$lambda.min, '\n 1Sd Lambda: ', cv.lasso$lambda.1se)
#> Min Lambda: 0.0224
#> 1Sd Lambda: 0.144
df_coef <- round(as.matrix(coef(cv.lasso, s=cv.lasso$lambda.min)), 2)
# See all contributing variables
df_coef[df_coef[, 1] != 0, ]
#> (Intercept) as mhci phci hvc vast
#> 2.68 -1.59 3.85 5.60 -2.41 -13.90
#> vars vari mdn mdi tmg tms
#> -20.18 -1.58 0.50 0.99 0.06 2.56
#> tmi
#> 2.23The above output shows what variables LASSO considered important. A high positive or low negative implies more important is that variable.
8.6 6. Step wise Forward and Backward Selection
Stepwise regression can be used to select features if the Y variable is a numeric variable. It is particularly used in selecting best linear regression models.
It searches for the best possible regression model by iteratively selecting and dropping variables to arrive at a model with the lowest possible AIC.
It can be implemented using the step() function and you need to
provide it with a lower model, which is the base model from which it
won’t remove any features and an upper model, which is a full model that
has all possible features you want to have.
Our case is not so complicated (< 20 vars), so lets just do a simple stepwise in ‘both’ directions.
I will use the ozone dataset for this where the objective is to
predict the ozone_reading based on other weather related observations.
# Load data
# online
# trainData <- read.csv("http://rstatistics.net/wp-content/uploads/2015/09/ozone1.csv",
# stringsAsFactors=F)
trainData <- read.csv(file.path(data_raw_dir, "ozone1.csv"))
print(head(trainData))
#> Month Day_of_month Day_of_week ozone_reading pressure_height Wind_speed
#> 1 1 1 4 3 5480 8
#> 2 1 2 5 3 5660 6
#> 3 1 3 6 3 5710 4
#> 4 1 4 7 5 5700 3
#> 5 1 5 1 5 5760 3
#> 6 1 6 2 6 5720 4
#> Humidity Temperature_Sandburg Temperature_ElMonte Inversion_base_height
#> 1 20 40.5 39.8 5000
#> 2 41 38.0 46.7 4109
#> 3 28 40.0 49.5 2693
#> 4 37 45.0 52.3 590
#> 5 51 54.0 45.3 1450
#> 6 69 35.0 49.6 1568
#> Pressure_gradient Inversion_temperature Visibility
#> 1 -15 30.6 200
#> 2 -14 48.0 300
#> 3 -25 47.7 250
#> 4 -24 55.0 100
#> 5 25 57.0 60
#> 6 15 53.8 60The data is ready. Let’s perform the stepwise.
# Step 1: Define base intercept only model
base.mod <- lm(ozone_reading ~ 1 , data=trainData)
# Step 2: Full model with all predictors
all.mod <- lm(ozone_reading ~ . , data= trainData)
# Step 3: Perform step-wise algorithm. direction='both' implies both forward and backward stepwise
stepMod <- step(base.mod, scope = list(lower = base.mod, upper = all.mod), direction = "both", trace = 0, steps = 1000)
# Step 4: Get the shortlisted variable.
shortlistedVars <- names(unlist(stepMod[[1]]))
shortlistedVars <- shortlistedVars[!shortlistedVars %in% "(Intercept)"] # remove intercept
# Show
print(shortlistedVars)
#> [1] "Temperature_Sandburg" "Humidity" "Temperature_ElMonte"
#> [4] "Month" "pressure_height" "Inversion_base_height"The selected model has the above 6 features in it.
But if you have too many features (> 100) in training data, then it might be a good idea to split the dataset into chunks of 10 variables each with Y as mandatory in each dataset. Loop through all the chunks and collect the best features.
We are doing it this way because some variables that came as important in a training data with fewer features may not show up in a linear reg model built on lots of features.
Finally, from a pool of shortlisted features (from small chunk models), run a full stepwise model to get the final set of selected features.
You can take this as a learning assignment to be solved within 20 minutes.
8.7 7. Relative Importance from Linear Regression
This technique is specific to linear regression models.
Relative importance can be used to assess which variables contributed how much in explaining the linear model’s R-squared value. So, if you sum up the produced importances, it will add up to the model’s R-sq value.
In essence, it is not directly a feature selection method, because you
have already provided the features that go in the model. But after
building the model, the relaimpo can provide a sense of how important
each feature is in contributing to the R-sq, or in other words, in
‘explaining the Y variable’.
So, how to calculate relative importance?
It is implemented in the relaimpo package. Basically, you build a
linear regression model and pass that as the main argument to
calc.relimp(). relaimpo has multiple options to compute the relative
importance, but the recommended method is to use type='lmg', as I have
done below.
# install.packages('relaimpo')
library(relaimpo)
#> Loading required package: boot
#>
#> Attaching package: 'boot'
#> The following object is masked from 'package:lattice':
#>
#> melanoma
#> The following object is masked from 'package:survival':
#>
#> aml
#> Loading required package: survey
#> Loading required package: grid
#>
#> Attaching package: 'survey'
#> The following object is masked from 'package:graphics':
#>
#> dotchart
#> Loading required package: mitools
#> This is the global version of package relaimpo.
#> If you are a non-US user, a version with the interesting additional metric pmvd is available
#> from Ulrike Groempings web site at prof.beuth-hochschule.de/groemping.
# Build linear regression model
model_formula = ozone_reading ~ Temperature_Sandburg + Humidity + Temperature_ElMonte + Month + pressure_height + Inversion_base_height
lmMod <- lm(model_formula, data=trainData)
# calculate relative importance
relImportance <- calc.relimp(lmMod, type = "lmg", rela = F)
# Sort
cat('Relative Importances: \n')
#> Relative Importances:
sort(round(relImportance$lmg, 3), decreasing=TRUE)
#> Temperature_ElMonte Temperature_Sandburg pressure_height
#> 0.214 0.203 0.104
#> Inversion_base_height Humidity Month
#> 0.096 0.086 0.012Additionally, you can use bootstrapping (using boot.relimp) to compute
the confidence intervals of the produced relative importances.
bootsub <- boot.relimp(ozone_reading ~ Temperature_Sandburg + Humidity + Temperature_ElMonte + Month + pressure_height + Inversion_base_height, data=trainData,
b = 1000, type = 'lmg', rank = TRUE, diff = TRUE)
plot(booteval.relimp(bootsub, level=.95))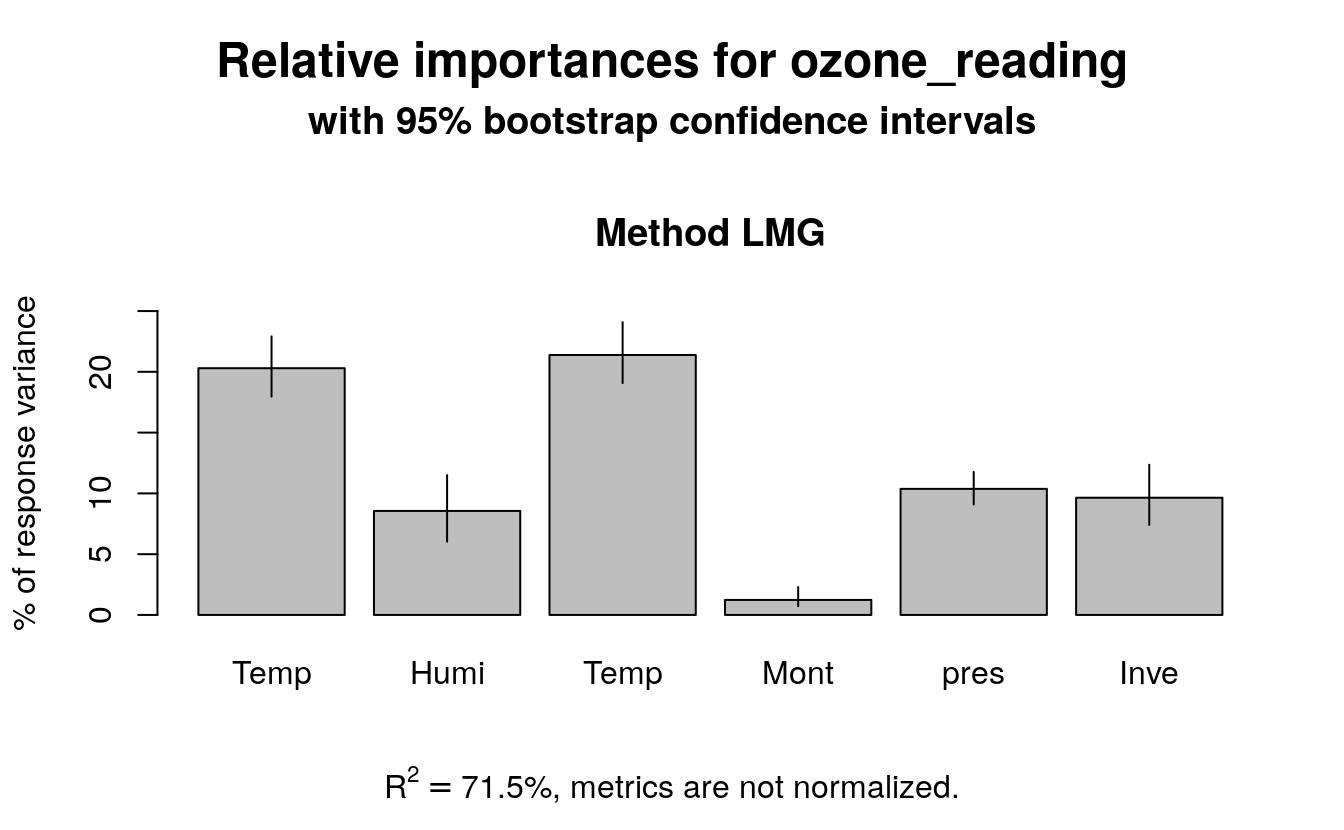
8.8 8. Recursive Feature Elimination (RFE)
Recursive feature elimnation (rfe) offers a rigorous way to determine the important variables before you even feed them into a ML algo.
It can be implemented using the rfe() from caret package.
The rfe() also takes two important parameters.
sizesrfeControl
So, what does sizes and rfeControl represent?
The sizes determines the number of most important features the rfe should iterate. Below, I have set the size as 1 to 5, 10, 15 and 18.
Secondly, the rfeControl parameter receives the output of the
rfeControl(). You can set what type of variable evaluation algorithm
must be used. Here, I have used random forests based rfFuncs. The
method='repeatedCV' means it will do a repeated k-Fold cross
validation with repeats=5.
Once complete, you get the accuracy and kappa for each model size you provided. The final selected model subset size is marked with a * in the rightmost selected column.
str(trainData)
#> 'data.frame': 366 obs. of 13 variables:
#> $ Month : int 1 1 1 1 1 1 1 1 1 1 ...
#> $ Day_of_month : int 1 2 3 4 5 6 7 8 9 10 ...
#> $ Day_of_week : int 4 5 6 7 1 2 3 4 5 6 ...
#> $ ozone_reading : num 3 3 3 5 5 6 4 4 6 7 ...
#> $ pressure_height : num 5480 5660 5710 5700 5760 5720 5790 5790 5700 5700 ...
#> $ Wind_speed : int 8 6 4 3 3 4 6 3 3 3 ...
#> $ Humidity : num 20 41 28 37 51 ...
#> $ Temperature_Sandburg : num 40.5 38 40 45 54 ...
#> $ Temperature_ElMonte : num 39.8 46.7 49.5 52.3 45.3 ...
#> $ Inversion_base_height: num 5000 4109 2693 590 1450 ...
#> $ Pressure_gradient : num -15 -14 -25 -24 25 15 -33 -28 23 -2 ...
#> $ Inversion_temperature: num 30.6 48 47.7 55 57 ...
#> $ Visibility : int 200 300 250 100 60 60 100 250 120 120 ...
tic()
set.seed(100)
options(warn=-1)
subsets <- c(1:5, 10, 15, 18)
ctrl <- rfeControl(functions = rfFuncs,
method = "repeatedcv",
repeats = 5,
verbose = FALSE)
lmProfile <- rfe(x=trainData[, c(1:3, 5:13)], y=trainData$ozone_reading,
sizes = subsets,
rfeControl = ctrl)
toc()
#> 92.495 sec elapsed
lmProfile
#>
#> Recursive feature selection
#>
#> Outer resampling method: Cross-Validated (10 fold, repeated 5 times)
#>
#> Resampling performance over subset size:
#>
#> Variables RMSE Rsquared MAE RMSESD RsquaredSD MAESD Selected
#> 1 5.13 0.595 3.92 0.826 0.1275 0.586
#> 2 4.03 0.746 3.11 0.542 0.0743 0.416
#> 3 3.95 0.756 3.06 0.472 0.0670 0.380
#> 4 3.93 0.759 3.01 0.468 0.0683 0.361
#> 5 3.90 0.763 2.98 0.467 0.0659 0.350
#> 10 3.77 0.782 2.85 0.496 0.0734 0.393 *
#> 12 3.77 0.781 2.86 0.508 0.0756 0.401
#>
#> The top 5 variables (out of 10):
#> Temperature_ElMonte, Pressure_gradient, Temperature_Sandburg, Inversion_temperature, HumiditySo, it says, Temperature_ElMonte, Pressure_gradient, Temperature_Sandburg, Inversion_temperature, Humidity are the top 5 variables in that order.
And the best model size out of the provided models sizes (in subsets) is 10.
You can see all of the top 10 variables from ‘lmProfile$optVariables’
that was created using rfe function above.
8.9 9. Genetic Algorithm
You can perform a supervised feature selection with genetic algorithms
using the gafs(). This is quite resource expensive so consider
that before choosing the number of iterations (iters) and the number of
repeats in gafsControl().
tic()
# Define control function
ga_ctrl <- gafsControl(functions = rfGA, # another option is `caretGA`.
method = "cv",
repeats = 3)
# Genetic Algorithm feature selection
set.seed(100)
ga_obj <- gafs(x=trainData[, c(1:3, 5:13)],
y=trainData[, 4],
iters = 3, # normally much higher (100+)
gafsControl = ga_ctrl)
toc()
#> 654.344 sec elapsed
ga_obj
#>
#> Genetic Algorithm Feature Selection
#>
#> 366 samples
#> 12 predictors
#>
#> Maximum generations: 3
#> Population per generation: 50
#> Crossover probability: 0.8
#> Mutation probability: 0.1
#> Elitism: 0
#>
#> Internal performance values: RMSE, Rsquared
#> Subset selection driven to minimize internal RMSE
#>
#> External performance values: RMSE, Rsquared, MAE
#> Best iteration chose by minimizing external RMSE
#> External resampling method: Cross-Validated (10 fold)
#>
#> During resampling:
#> * the top 5 selected variables (out of a possible 12):
#> Month (100%), Pressure_gradient (100%), Temperature_ElMonte (100%), Humidity (80%), Visibility (80%)
#> * on average, 6.8 variables were selected (min = 5, max = 9)
#>
#> In the final search using the entire training set:
#> * 9 features selected at iteration 2 including:
#> Month, Day_of_month, pressure_height, Wind_speed, Humidity ...
#> * external performance at this iteration is
#>
#> RMSE Rsquared MAE
#> 3.721 0.788 2.800
# Optimal variables
ga_obj$optVariables
#> [1] "Month" "Day_of_month" "pressure_height"
#> [4] "Wind_speed" "Humidity" "Temperature_ElMonte"
#> [7] "Inversion_base_height" "Pressure_gradient" "Inversion_temperature"‘Month’ ‘Day_of_month’ ‘Wind_speed’ ‘Temperature_ElMonte’ ‘Pressure_gradient’ ‘Visibility’
So the optimal variables according to the genetic algorithms are listed above. But, I wouldn’t use it just yet because, the above variant was tuned for only 3 iterations, which is quite low. I had to set it so low to save computing time.
8.10 10. Simulated Annealing
Simulated annealing is a global search algorithm that allows a suboptimal solution to be accepted in hope that a better solution will show up eventually.
It works by making small random changes to an initial solution and sees if the performance improved. The change is accepted if it improves, else it can still be accepted if the difference of performances meet an acceptance criteria.
In caret it has been implemented in the safs() which accepts a control
parameter that can be set using the safsControl() function.
safsControl is similar to other control functions in caret (like you
saw in rfe and ga), and additionally it accepts an improve parameter
which is the number of iterations it should wait without improvement
until the values are reset to previous iteration.
tic()
# Define control function
sa_ctrl <- safsControl(functions = rfSA,
method = "repeatedcv",
repeats = 3,
improve = 5) # n iterations without improvement before a reset
# Genetic Algorithm feature selection
set.seed(100)
sa_obj <- safs(x=trainData[, c(1:3, 5:13)],
y=trainData[, 4],
safsControl = sa_ctrl)
toc()
#> 113.053 sec elapsed
sa_obj
#>
#> Simulated Annealing Feature Selection
#>
#> 366 samples
#> 12 predictors
#>
#> Maximum search iterations: 10
#> Restart after 5 iterations without improvement (0.3 restarts on average)
#>
#> Internal performance values: RMSE, Rsquared
#> Subset selection driven to minimize internal RMSE
#>
#> External performance values: RMSE, Rsquared, MAE
#> Best iteration chose by minimizing external RMSE
#> External resampling method: Cross-Validated (10 fold, repeated 3 times)
#>
#> During resampling:
#> * the top 5 selected variables (out of a possible 12):
#> Temperature_Sandburg (80%), Month (66.7%), Pressure_gradient (66.7%), Temperature_ElMonte (63.3%), Visibility (60%)
#> * on average, 6.5 variables were selected (min = 3, max = 11)
#>
#> In the final search using the entire training set:
#> * 6 features selected at iteration 9 including:
#> Day_of_week, pressure_height, Wind_speed, Humidity, Inversion_base_height ...
#> * external performance at this iteration is
#>
#> RMSE Rsquared MAE
#> 4.108 0.743 3.111
# Optimal variables
print(sa_obj$optVariables)
#> [1] "Day_of_week" "pressure_height" "Wind_speed"
#> [4] "Humidity" "Inversion_base_height" "Pressure_gradient"8.11 Information Value and Weights of Evidence
The Information Value can be used to judge how important a given categorical variable is in explaining the binary Y variable. It goes well with logistic regression and other classification models that can model binary variables.
Let’s try to find out how important the categorical variables are in
predicting if an individual will earn > 50k from the adult.csv
dataset. Just run the code below to import the dataset.
library(InformationValue)
#>
#> Attaching package: 'InformationValue'
#> The following objects are masked from 'package:caret':
#>
#> confusionMatrix, precision, sensitivity, specificity
# online data
# inputData <- read.csv("http://rstatistics.net/wp-content/uploads/2015/09/adult.csv")
inputData <- read.csv(file.path(data_raw_dir, "adult.csv"))
print(head(inputData))
#> AGE WORKCLASS FNLWGT EDUCATION EDUCATIONNUM MARITALSTATUS
#> 1 39 State-gov 77516 Bachelors 13 Never-married
#> 2 50 Self-emp-not-inc 83311 Bachelors 13 Married-civ-spouse
#> 3 38 Private 215646 HS-grad 9 Divorced
#> 4 53 Private 234721 11th 7 Married-civ-spouse
#> 5 28 Private 338409 Bachelors 13 Married-civ-spouse
#> 6 37 Private 284582 Masters 14 Married-civ-spouse
#> OCCUPATION RELATIONSHIP RACE SEX CAPITALGAIN CAPITALLOSS
#> 1 Adm-clerical Not-in-family White Male 2174 0
#> 2 Exec-managerial Husband White Male 0 0
#> 3 Handlers-cleaners Not-in-family White Male 0 0
#> 4 Handlers-cleaners Husband Black Male 0 0
#> 5 Prof-specialty Wife Black Female 0 0
#> 6 Exec-managerial Wife White Female 0 0
#> HOURSPERWEEK NATIVECOUNTRY ABOVE50K
#> 1 40 United-States 0
#> 2 13 United-States 0
#> 3 40 United-States 0
#> 4 40 United-States 0
#> 5 40 Cuba 0
#> 6 40 United-States 0
# Choose Categorical Variables to compute Info Value.
cat_vars <- c ("WORKCLASS", "EDUCATION", "MARITALSTATUS", "OCCUPATION", "RELATIONSHIP", "RACE", "SEX", "NATIVECOUNTRY") # get all categorical variables
factor_vars <- cat_vars
# Init Output
df_iv <- data.frame(VARS=cat_vars, IV=numeric(length(cat_vars)), STRENGTH=character(length(cat_vars)), stringsAsFactors = F) # init output dataframe
# Get Information Value for each variable
for (factor_var in factor_vars){
df_iv[df_iv$VARS == factor_var, "IV"] <- InformationValue::IV(X=inputData[, factor_var], Y=inputData$ABOVE50K)
df_iv[df_iv$VARS == factor_var, "STRENGTH"] <- attr(InformationValue::IV(X=inputData[, factor_var], Y=inputData$ABOVE50K), "howgood")
}
# Sort
df_iv <- df_iv[order(-df_iv$IV), ]
df_iv
#> VARS IV STRENGTH
#> 5 RELATIONSHIP 1.5356 Highly Predictive
#> 3 MARITALSTATUS 1.3388 Highly Predictive
#> 4 OCCUPATION 0.7762 Highly Predictive
#> 2 EDUCATION 0.7411 Highly Predictive
#> 7 SEX 0.3033 Highly Predictive
#> 1 WORKCLASS 0.1634 Highly Predictive
#> 8 NATIVECOUNTRY 0.0794 Somewhat Predictive
#> 6 RACE 0.0693 Somewhat PredictiveHere is what the quantum of Information Value means:
Less than 0.02, then the predictor is not useful for modeling (separating the Goods from the Bads)
0.02 to 0.1, then the predictor has only a weak relationship. 0.1 to 0.3, then the predictor has a medium strength relationship. 0.3 or higher, then the predictor has a strong relationship. That was about IV. Then what is Weight of Evidence?
Weights of evidence can be useful to find out how important a given categorical variable is in explaining the ‘events’ (called ‘Goods’ in below table.)
The ‘Information Value’ of the categorical variable can then be derived from the respective WOE values.
IV=(perc good of all goods−perc bad of all bads) *WOE
The ‘WOETable’ below given the computation in more detail.
WOETable(X=inputData[, 'WORKCLASS'], Y=inputData$ABOVE50K)
#> CAT GOODS BADS TOTAL PCT_G PCT_B WOE IV
#> 1 ? 191 1645 1836 0.02429 0.066545 -1.008 0.042574
#> 2 Federal-gov 371 589 960 0.04719 0.023827 0.683 0.015964
#> 3 Local-gov 617 1476 2093 0.07848 0.059709 0.273 0.005131
#> 4 Never-worked 7 7 7 0.00089 0.000283 1.146 0.000696
#> 5 Private 4963 17733 22696 0.63126 0.717354 -0.128 0.011006
#> 6 Self-emp-inc 622 494 1116 0.07911 0.019984 1.376 0.081363
#> 7 Self-emp-not-inc 724 1817 2541 0.09209 0.073503 0.225 0.004190
#> 8 State-gov 353 945 1298 0.04490 0.038228 0.161 0.001073
#> 9 Without-pay 14 14 14 0.00178 0.000566 1.146 0.001391The total IV of a variable is the sum of IV’s of its categories.
8.12 DALEX Package
The DALEX is a powerful package that explains various things about the
variables used in an ML model.
For example, using the variable_dropout() function you can find out
how important a variable is based on a dropout loss, that is how much
loss is incurred by removing a variable from the model.
Apart from this, it also has the single_variable() function that gives
you an idea of how the model’s output will change by changing the values
of one of the X’s in the model.
It also has the single_prediction() that can decompose a single model
prediction so as to understand which variable caused what effect in
predicting the value of Y.
library(randomForest)
#> randomForest 4.6-14
#> Type rfNews() to see new features/changes/bug fixes.
#>
#> Attaching package: 'randomForest'
#> The following object is masked from 'package:dplyr':
#>
#> combine
#> The following object is masked from 'package:ranger':
#>
#> importance
#> The following object is masked from 'package:ggplot2':
#>
#> margin
library(DALEX)
# Load data
# inputData <- read.csv("http://rstatistics.net/wp-content/uploads/2015/09/adult.csv")
inputData <- read.csv(file.path(data_raw_dir, "adult.csv"))
# Train random forest model
rf_mod <- randomForest(factor(ABOVE50K) ~ ., data = inputData, ntree=100)
rf_mod
#>
#> Call:
#> randomForest(formula = factor(ABOVE50K) ~ ., data = inputData, ntree = 100)
#> Type of random forest: classification
#> Number of trees: 100
#> No. of variables tried at each split: 3
#>
#> OOB estimate of error rate: 13.6%
#> Confusion matrix:
#> 0 1 class.error
#> 0 23051 1669 0.0675
#> 1 2754 5087 0.3512
# Variable importance with DALEX
explained_rf <- explain(rf_mod, data=inputData, y=inputData$ABOVE50K)
#> Preparation of a new explainer is initiated
#> -> model label : randomForest ( default )
#> -> data : 32561 rows 15 cols
#> -> target variable : 32561 values
#> -> model_info : package randomForest , ver. 4.6.14 , task classification ( default )
#> -> predict function : yhat.randomForest will be used ( default )
#> -> predicted values : numerical, min = 0 , mean = 0.237 , max = 1
#> -> residual function : difference between y and yhat ( default )
#> -> residuals : numerical, min = -0.94 , mean = 0.00374 , max = 0.93
#> A new explainer has been created!
# Get the variable importances
# varimps = variable_dropout(explained_rf, type='raw')
varimps = variable_importance(explained_rf, type='raw')
print(varimps)
#> variable mean_dropout_loss label
#> 1 _full_model_ 36.8 randomForest
#> 2 ABOVE50K 36.8 randomForest
#> 3 RACE 41.1 randomForest
#> 4 NATIVECOUNTRY 43.6 randomForest
#> 5 SEX 46.4 randomForest
#> 6 CAPITALLOSS 46.9 randomForest
#> 7 WORKCLASS 55.8 randomForest
#> 8 EDUCATION 62.1 randomForest
#> 9 EDUCATIONNUM 63.3 randomForest
#> 10 FNLWGT 64.0 randomForest
#> 11 RELATIONSHIP 64.9 randomForest
#> 12 HOURSPERWEEK 68.6 randomForest
#> 13 CAPITALGAIN 69.3 randomForest
#> 14 MARITALSTATUS 73.3 randomForest
#> 15 OCCUPATION 85.1 randomForest
#> 16 AGE 85.2 randomForest
#> 17 _baseline_ 299.5 randomForest
plot(varimps)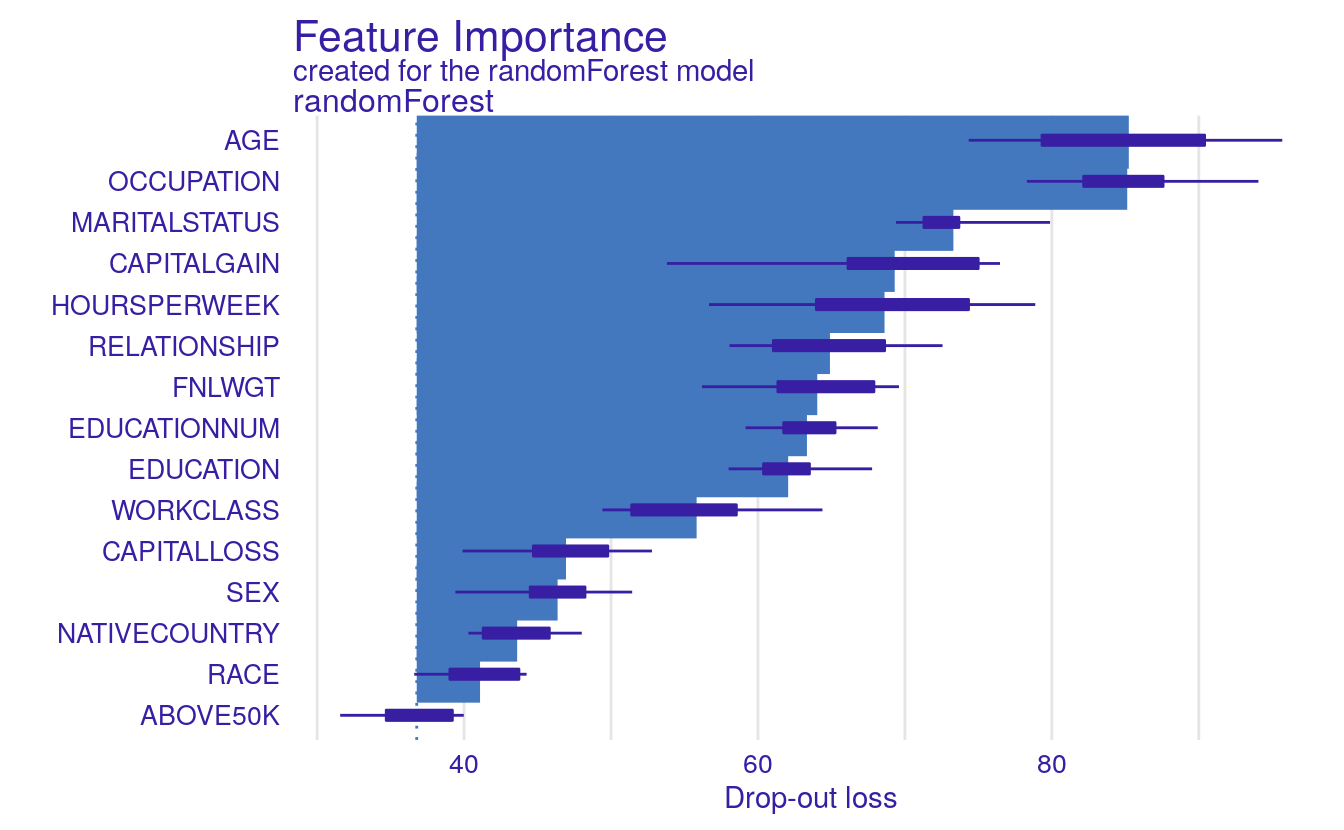
8.13 Conclusion
Hope you find these methods useful. As it turns out different methods
showed different variables as important, or at least the degree of
importance changed. This need not be a conflict, because each method
gives a different perspective of how the variable can be useful
depending on how the algorithms learn Y ~ x. So its cool.
If you find any code breaks or bugs, report the issue here or just write it below.