38 Temperature modeling using nested dataframes
Dataset: temperature.csv
38.1 Introduction
http://ijlyttle.github.io/isugg_purrr/presentation.html#(1)
38.1.2 Motivation
As you know, purrr is a recent package from Hadley Wickham, focused on lists and functional programming, like dplyr is focused on data-frames.
I figure a good way to learn a new package is to try to solve a problem, so we have a dataset:
you can download the source of this presentation
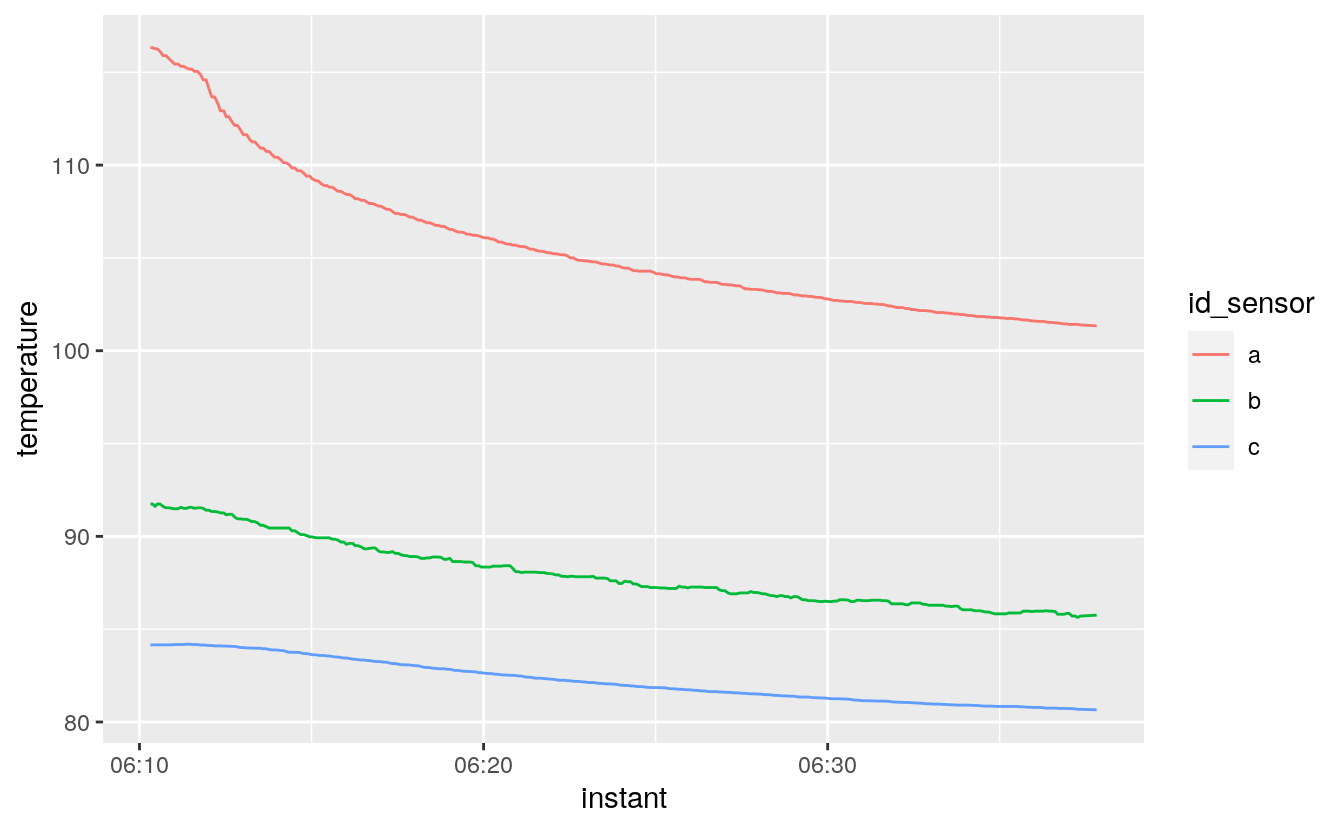
these are three temperatures recorded simultaneously in a piece of electronics
it will be very valuable to be able to characterize the transient temperature for each sensor
we want to apply the same set of models across all three sensors
it will be easier to show using pictures
38.2 Prepare the data
38.2.1 Let’s get the data into shape
Using the readr package
temperature_wide <-
read_csv(file.path(data_raw_dir, "temperature.csv")) %>%
print()
#> Parsed with column specification:
#> cols(
#> instant = col_datetime(format = ""),
#> temperature_a = col_double(),
#> temperature_b = col_double(),
#> temperature_c = col_double()
#> )
#> # A tibble: 327 x 4
#> instant temperature_a temperature_b temperature_c
#> <dttm> <dbl> <dbl> <dbl>
#> 1 2015-11-13 06:10:19 116. 91.7 84.2
#> 2 2015-11-13 06:10:23 116. 91.7 84.2
#> 3 2015-11-13 06:10:27 116. 91.6 84.2
#> 4 2015-11-13 06:10:31 116. 91.7 84.2
#> 5 2015-11-13 06:10:36 116. 91.7 84.2
#> 6 2015-11-13 06:10:41 116. 91.6 84.2
#> # … with 321 more rows
38.2.2 Is temperature_wide “tidy”?
#> # A tibble: 327 x 4
#> instant temperature_a temperature_b temperature_c
#> <dttm> <dbl> <dbl> <dbl>
#> 1 2015-11-13 06:10:19 116. 91.7 84.2
#> 2 2015-11-13 06:10:23 116. 91.7 84.2
#> 3 2015-11-13 06:10:27 116. 91.6 84.2
#> 4 2015-11-13 06:10:31 116. 91.7 84.2
#> 5 2015-11-13 06:10:36 116. 91.7 84.2
#> 6 2015-11-13 06:10:41 116. 91.6 84.2
#> # … with 321 more rowsWhy or why not?
38.2.3 Tidy data
- Each column is a variable
- Each row is an observation
- Each cell is a value
(http://www.jstatsoft.org/v59/i10/paper)
My personal observation is that “tidy” can depend on the context, on what you want to do with the data.
38.2.4 Let’s get this into a tidy form
temperature_tall <-
temperature_wide %>%
gather(key = "id_sensor", value = "temperature", starts_with("temp")) %>%
mutate(id_sensor = str_replace(id_sensor, "temperature_", "")) %>%
print()
#> # A tibble: 981 x 3
#> instant id_sensor temperature
#> <dttm> <chr> <dbl>
#> 1 2015-11-13 06:10:19 a 116.
#> 2 2015-11-13 06:10:23 a 116.
#> 3 2015-11-13 06:10:27 a 116.
#> 4 2015-11-13 06:10:31 a 116.
#> 5 2015-11-13 06:10:36 a 116.
#> 6 2015-11-13 06:10:41 a 116.
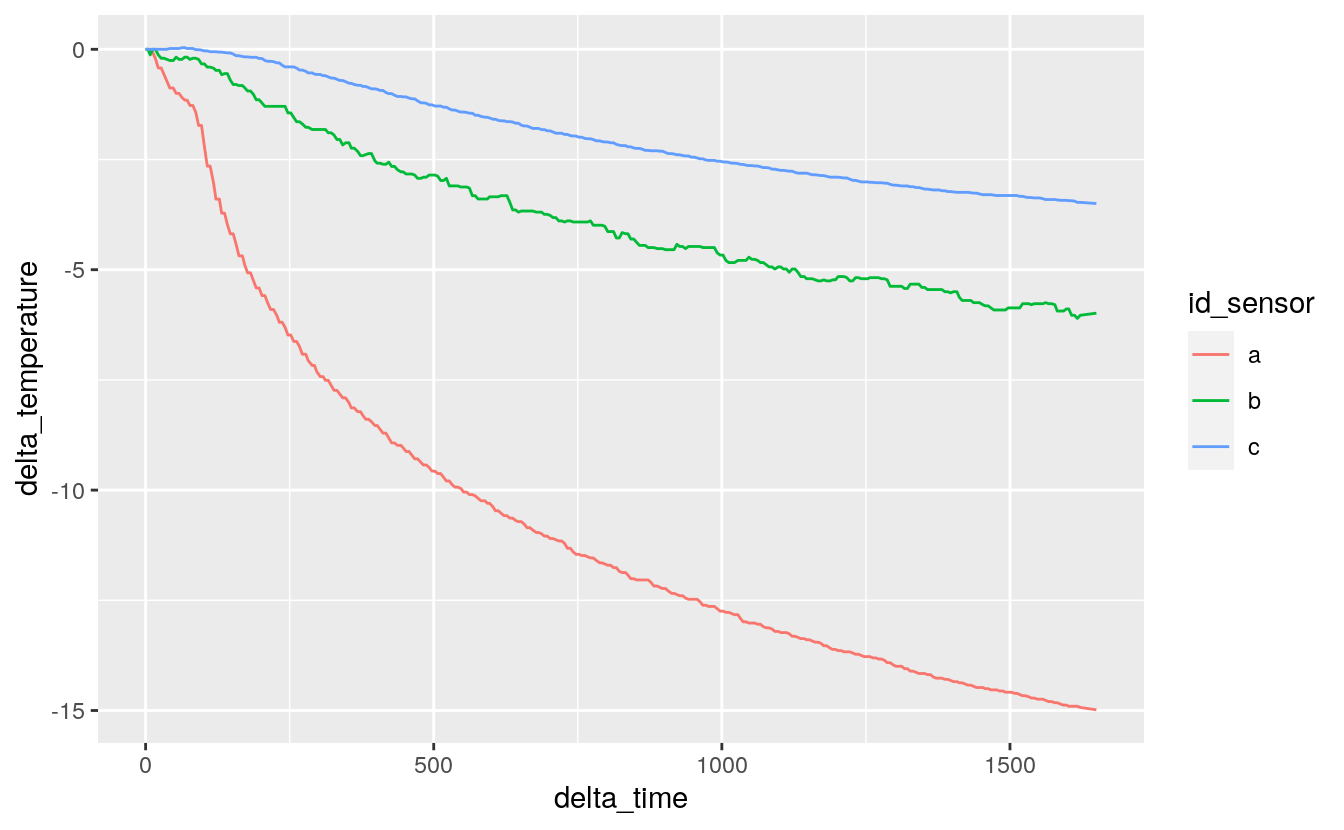
#> # … with 975 more rows38.2.6 Calculate delta time (\(\Delta t\)) and delta temperature (\(\Delta T\))
delta_time \(\Delta t\)
change in time since event started, s
delta_temperature: \(\Delta T\)
change in temperature since event started, °C
delta <-
temperature_tall %>%
arrange(id_sensor, instant) %>%
group_by(id_sensor) %>%
mutate(
delta_time = as.numeric(instant) - as.numeric(instant[[1]]),
delta_temperature = temperature - temperature[[1]]
) %>%
select(id_sensor, delta_time, delta_temperature)38.3 Define the models
We want to see how three different curve-fits might perform on these three data-sets:
38.3.2 Semi-infinite solid with convection
\[\Delta T = \Delta T_0 * \big [ \operatorname erfc(\sqrt{\frac{\tau_0}{\delta t}}) - e^ {Bi_0 + (\frac {Bi_0}{2})^2 \frac {\delta t}{\tau_0}} * \operatorname erfc (\sqrt \frac{\tau_0}{\delta t} + \frac {Bi_0}{2} * \sqrt \frac{\delta t }{\tau_0} \big]\]
38.3.5 Temperature models: simple and convection
semi_infinite_simple <- function(x) {
nls(
delta_temperature ~ delta_temperature_0 * erfc(sqrt(tau_0 / delta_time)),
start = list(delta_temperature_0 = -10, tau_0 = 50),
data = x
)
}
semi_infinite_convection <- function(x){
nls(
delta_temperature ~
delta_temperature_0 * (
erfc(sqrt(tau_0 / delta_time)) -
exp(Bi_0 + (Bi_0/2)^2 * delta_time / tau_0) *
erfc(sqrt(tau_0 / delta_time) +
(Bi_0/2) * sqrt(delta_time / tau_0))
),
start = list(delta_temperature_0 = -5, tau_0 = 50, Bi_0 = 1.e6),
data = x
)
}38.4 Test modeling on one dataset
38.4.1 Before going into purrr
Before doing anything, we want to show that we can do something with one dataset and one model-function:
# only one sensor; it is a test
tmp_data <- delta %>% filter(id_sensor == "a")
tmp_model <- newton_cooling(tmp_data)
summary(tmp_model)
#>
#> Formula: delta_temperature ~ delta_temperature_0 * (1 - exp(-delta_time/tau_0))
#>
#> Parameters:
#> Estimate Std. Error t value Pr(>|t|)
#> delta_temperature_0 -15.0608 0.0526 -286 <2e-16 ***
#> tau_0 500.0138 4.8367 103 <2e-16 ***
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> Residual standard error: 0.327 on 325 degrees of freedom
#>
#> Number of iterations to convergence: 7
#> Achieved convergence tolerance: 4.14e-0638.4.2 Look at predictions
# apply prediction and make it tidy
tmp_pred <-
tmp_data %>%
mutate(modeled = predict(tmp_model, data = .)) %>%
select(id_sensor, delta_time, measured = delta_temperature, modeled) %>%
gather("type", "delta_temperature", measured:modeled) %>%
print()
#> # A tibble: 654 x 4
#> # Groups: id_sensor [1]
#> id_sensor delta_time type delta_temperature
#> <chr> <dbl> <chr> <dbl>
#> 1 a 0 measured 0
#> 2 a 4 measured 0
#> 3 a 8 measured -0.06
#> 4 a 12 measured -0.06
#> 5 a 17 measured -0.211
#> 6 a 22 measured -0.423
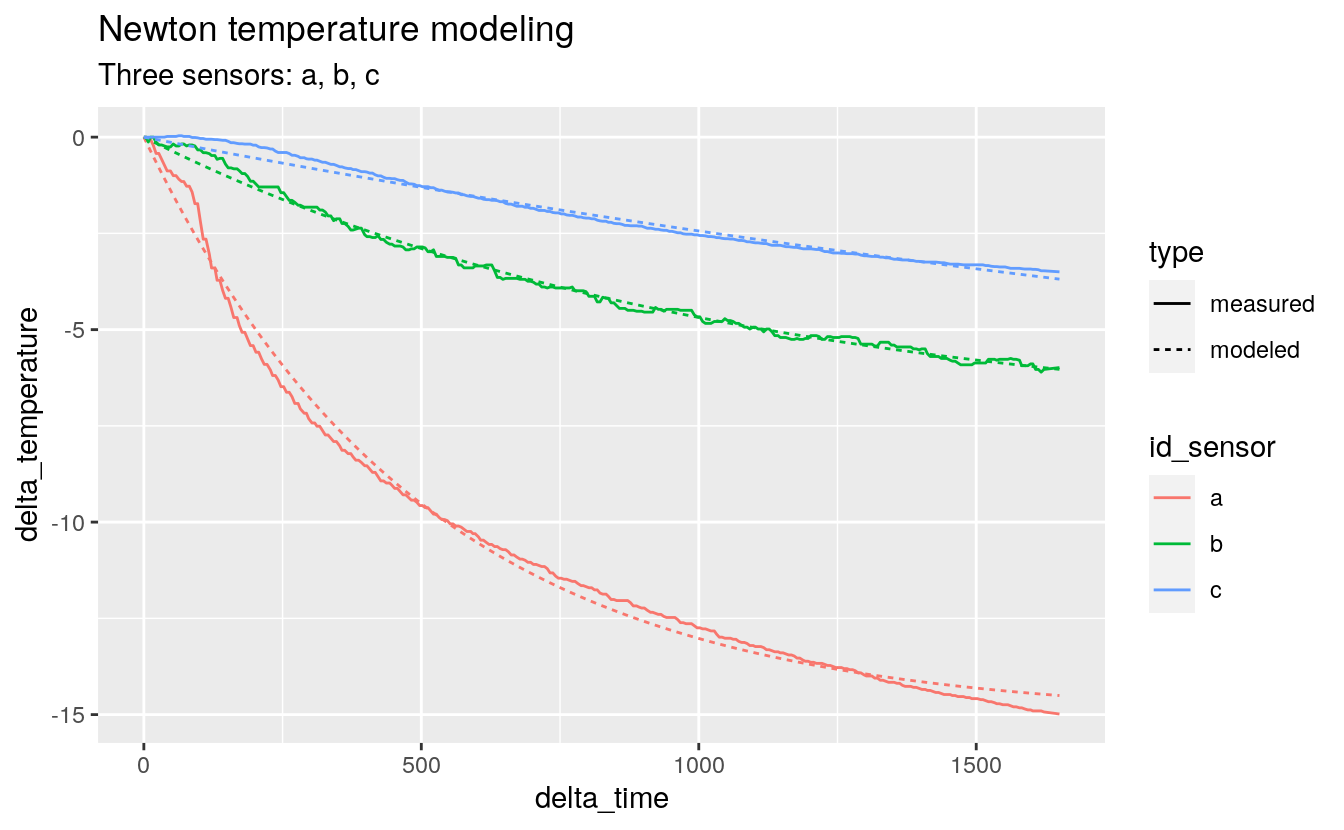
#> # … with 648 more rows38.4.3 Plot Newton model
tmp_pred %>%
ggplot(aes(x = delta_time, y = delta_temperature, linetype = type)) +
geom_line() +
labs(title = "Newton temperature model", subtitle = "One sensor: a")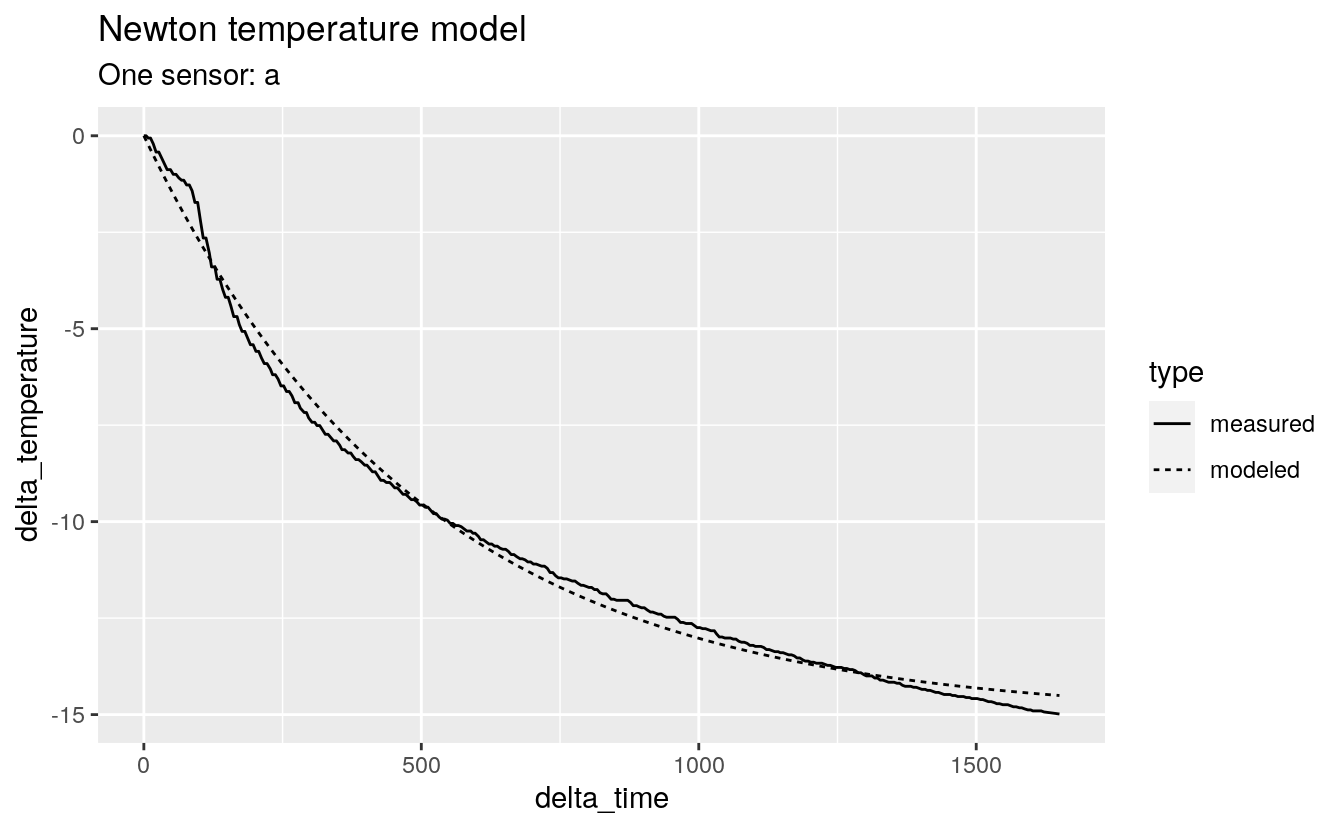
38.4.4 “Regular” data-frame (deltas)
print(delta)
#> # A tibble: 981 x 3
#> # Groups: id_sensor [3]
#> id_sensor delta_time delta_temperature
#> <chr> <dbl> <dbl>
#> 1 a 0 0
#> 2 a 4 0
#> 3 a 8 -0.06
#> 4 a 12 -0.06
#> 5 a 17 -0.211
#> 6 a 22 -0.423
#> # … with 975 more rowsEach column of the dataframe is a vector - in this case, a character vector and two doubles
38.5 Making a nested dataframe
38.5.1 How to make a weird data-frame
Here’s where the fun starts - a column of a data-frame can be a list.
use
tidyr::nest()to makes a columndata, which is a list of data-framesthis seems like a stronger expression of the
dplyr::group_by()idea
# nest delta_time and delta_temperature variables
delta_nested <-
delta %>%
nest(-id_sensor) %>%
print()
#> Warning: All elements of `...` must be named.
#> Did you want `data = c(delta_time, delta_temperature)`?
#> # A tibble: 3 x 2
#> # Groups: id_sensor [3]
#> id_sensor data
#> <chr> <list>
#> 1 a <tibble [327 × 2]>
#> 2 b <tibble [327 × 2]>
#> 3 c <tibble [327 × 2]>38.5.2 Map dataframes to a modeling function (Newton)
model_nested <-
delta_nested %>%
mutate(model = map(data, newton_cooling)) %>%
print()
#> # A tibble: 3 x 3
#> # Groups: id_sensor [3]
#> id_sensor data model
#> <chr> <list> <list>
#> 1 a <tibble [327 × 2]> <nls>
#> 2 b <tibble [327 × 2]> <nls>
#> 3 c <tibble [327 × 2]> <nls>We get an additional list-column
model.
38.5.3 We can use map2() to make the predictions
predict_nested <-
model_nested %>%
mutate(pred = map2(model, data, predict)) %>%
print()
#> # A tibble: 3 x 4
#> # Groups: id_sensor [3]
#> id_sensor data model pred
#> <chr> <list> <list> <list>
#> 1 a <tibble [327 × 2]> <nls> <dbl [327]>
#> 2 b <tibble [327 × 2]> <nls> <dbl [327]>
#> 3 c <tibble [327 × 2]> <nls> <dbl [327]>Another list-column
predfor the prediction results.
38.5.4 We need to get out of the weirdness
- use
unnest()to get back to a regular data-frame
predict_unnested <-
predict_nested %>%
unnest(data, pred) %>%
print()
#> Warning: unnest() has a new interface. See ?unnest for details.
#> Try `df %>% unnest(c(data, pred))`, with `mutate()` if needed
#> # A tibble: 981 x 5
#> # Groups: id_sensor [3]
#> id_sensor delta_time delta_temperature model pred
#> <chr> <dbl> <dbl> <list> <dbl>
#> 1 a 0 0 <nls> 0
#> 2 a 4 0 <nls> -0.120
#> 3 a 8 -0.06 <nls> -0.239
#> 4 a 12 -0.06 <nls> -0.357
#> 5 a 17 -0.211 <nls> -0.503
#> 6 a 22 -0.423 <nls> -0.648
#> # … with 975 more rows38.5.5 We can wrangle the predictions
- get into a form that makes it easier to plot
predict_tall <-
predict_unnested %>%
rename(modeled = pred, measured = delta_temperature) %>%
gather("type", "delta_temperature", modeled, measured) %>%
print()
#> # A tibble: 1,962 x 5
#> # Groups: id_sensor [3]
#> id_sensor delta_time model type delta_temperature
#> <chr> <dbl> <list> <chr> <dbl>
#> 1 a 0 <nls> modeled 0
#> 2 a 4 <nls> modeled -0.120
#> 3 a 8 <nls> modeled -0.239
#> 4 a 12 <nls> modeled -0.357
#> 5 a 17 <nls> modeled -0.503
#> 6 a 22 <nls> modeled -0.648
#> # … with 1,956 more rows38.6 Apply multiple models on a nested structure
38.6.1 Step 1: Selection of models
Make a list of functions to model:
list_model <-
list(
newton_cooling = newton_cooling,
semi_infinite_simple = semi_infinite_simple,
semi_infinite_convection = semi_infinite_convection
)38.6.2 Step 2: write a function to define the “inner” loop
# add additional variable with the model name
fn_model <- function(.model, df) {
# one parameter for the model in the list, the second for the data
# safer to avoid non-standard evaluation
# df %>% mutate(model = map(data, .model))
df$model <- map(df$data, possibly(.model, NULL))
df
}for a given model-function and a given (weird) data-frame, return a modified version of that data-frame with a column
model, which is the model-function applied to each element of the data-frame’sdatacolumn (which is itself a list of data-frames)the purrr functions
safely()andpossibly()are very interesting. I think they could be useful outside of purrr as a friendlier way to do error-handling.
38.6.3 Step 3: Use map_df() to define the “outer” loop
# this dataframe will be the second input of fn_model
delta_nested %>%
print()
#> # A tibble: 3 x 2
#> # Groups: id_sensor [3]
#> id_sensor data
#> <chr> <list>
#> 1 a <tibble [327 × 2]>
#> 2 b <tibble [327 × 2]>
#> 3 c <tibble [327 × 2]>
# fn_model is receiving two inputs: one from list_model and from delta_nested
model_nested_new <-
list_model %>%
map_df(fn_model, delta_nested, .id = "id_model") %>%
print()
#> # A tibble: 9 x 4
#> # Groups: id_sensor [3]
#> id_model id_sensor data model
#> <chr> <chr> <list> <list>
#> 1 newton_cooling a <tibble [327 × 2]> <nls>
#> 2 newton_cooling b <tibble [327 × 2]> <nls>
#> 3 newton_cooling c <tibble [327 × 2]> <nls>
#> 4 semi_infinite_simple a <tibble [327 × 2]> <nls>
#> 5 semi_infinite_simple b <tibble [327 × 2]> <nls>
#> 6 semi_infinite_simple c <tibble [327 × 2]> <nls>
#> # … with 3 more rowsfor each element of a list of model-functions, run the inner-loop function, and row-bind the results into a data-frame
we want to discard the rows where the model failed
we also want to investigate why they failed, but that’s a different talk
38.6.4 Step 4: Use map() to identify the null models
model_nested_new <-
list_model %>%
map_df(fn_model, delta_nested, .id = "id_model") %>%
mutate(is_null = map(model, is.null)) %>%
print()
#> # A tibble: 9 x 5
#> # Groups: id_sensor [3]
#> id_model id_sensor data model is_null
#> <chr> <chr> <list> <list> <list>
#> 1 newton_cooling a <tibble [327 × 2]> <nls> <lgl [1]>
#> 2 newton_cooling b <tibble [327 × 2]> <nls> <lgl [1]>
#> 3 newton_cooling c <tibble [327 × 2]> <nls> <lgl [1]>
#> 4 semi_infinite_simple a <tibble [327 × 2]> <nls> <lgl [1]>
#> 5 semi_infinite_simple b <tibble [327 × 2]> <nls> <lgl [1]>
#> 6 semi_infinite_simple c <tibble [327 × 2]> <nls> <lgl [1]>
#> # … with 3 more rows- using
map(model, is.null)returns a list column - to use
filter(), we have to escape the weirdness
38.6.5 Step 5: map_lgl() to identify nulls and get out of the weirdness
model_nested_new <-
list_model %>%
map_df(fn_model, delta_nested, .id = "id_model") %>%
mutate(is_null = map_lgl(model, is.null)) %>%
print()
#> # A tibble: 9 x 5
#> # Groups: id_sensor [3]
#> id_model id_sensor data model is_null
#> <chr> <chr> <list> <list> <lgl>
#> 1 newton_cooling a <tibble [327 × 2]> <nls> FALSE
#> 2 newton_cooling b <tibble [327 × 2]> <nls> FALSE
#> 3 newton_cooling c <tibble [327 × 2]> <nls> FALSE
#> 4 semi_infinite_simple a <tibble [327 × 2]> <nls> FALSE
#> 5 semi_infinite_simple b <tibble [327 × 2]> <nls> FALSE
#> 6 semi_infinite_simple c <tibble [327 × 2]> <nls> FALSE
#> # … with 3 more rows- using
map_lgl(model, is.null)returns a vector column
38.6.6 Step 6: filter() nulls and select() variables to clean up
model_nested_new <-
list_model %>%
map_df(fn_model, delta_nested, .id = "id_model") %>%
mutate(is_null = map_lgl(model, is.null)) %>%
filter(!is_null) %>%
select(-is_null) %>%
print()
#> # A tibble: 6 x 4
#> # Groups: id_sensor [3]
#> id_model id_sensor data model
#> <chr> <chr> <list> <list>
#> 1 newton_cooling a <tibble [327 × 2]> <nls>
#> 2 newton_cooling b <tibble [327 × 2]> <nls>
#> 3 newton_cooling c <tibble [327 × 2]> <nls>
#> 4 semi_infinite_simple a <tibble [327 × 2]> <nls>
#> 5 semi_infinite_simple b <tibble [327 × 2]> <nls>
#> 6 semi_infinite_simple c <tibble [327 × 2]> <nls>38.6.7 Step 7: Calculate predictions on nested dataframe
predict_nested <-
model_nested_new %>%
mutate(pred = map2(model, data, predict)) %>%
print()
#> # A tibble: 6 x 5
#> # Groups: id_sensor [3]
#> id_model id_sensor data model pred
#> <chr> <chr> <list> <list> <list>
#> 1 newton_cooling a <tibble [327 × 2]> <nls> <dbl [327]>
#> 2 newton_cooling b <tibble [327 × 2]> <nls> <dbl [327]>
#> 3 newton_cooling c <tibble [327 × 2]> <nls> <dbl [327]>
#> 4 semi_infinite_simple a <tibble [327 × 2]> <nls> <dbl [327]>
#> 5 semi_infinite_simple b <tibble [327 × 2]> <nls> <dbl [327]>
#> 6 semi_infinite_simple c <tibble [327 × 2]> <nls> <dbl [327]>
38.6.8 unnest(), make it tall and tidy
predict_tall <-
predict_nested %>%
unnest(data, pred) %>%
rename(modeled = pred, measured = delta_temperature) %>%
gather("type", "delta_temperature", modeled, measured) %>%
print()
#> Warning: unnest() has a new interface. See ?unnest for details.
#> Try `df %>% unnest(c(data, pred))`, with `mutate()` if needed
#> # A tibble: 3,924 x 6
#> # Groups: id_sensor [3]
#> id_model id_sensor delta_time model type delta_temperature
#> <chr> <chr> <dbl> <list> <chr> <dbl>
#> 1 newton_cooling a 0 <nls> modeled 0
#> 2 newton_cooling a 4 <nls> modeled -0.120
#> 3 newton_cooling a 8 <nls> modeled -0.239
#> 4 newton_cooling a 12 <nls> modeled -0.357
#> 5 newton_cooling a 17 <nls> modeled -0.503
#> 6 newton_cooling a 22 <nls> modeled -0.648
#> # … with 3,918 more rows38.6.9 Visualize the predictions
predict_tall %>%
ggplot(aes(x = delta_time, y = delta_temperature)) +
geom_line(aes(color = id_sensor, linetype = type)) +
facet_grid(id_model ~ .) +
labs(title = "Newton and Semi-infinite temperature modeling",
subtitle = "Three sensors: a, b, c")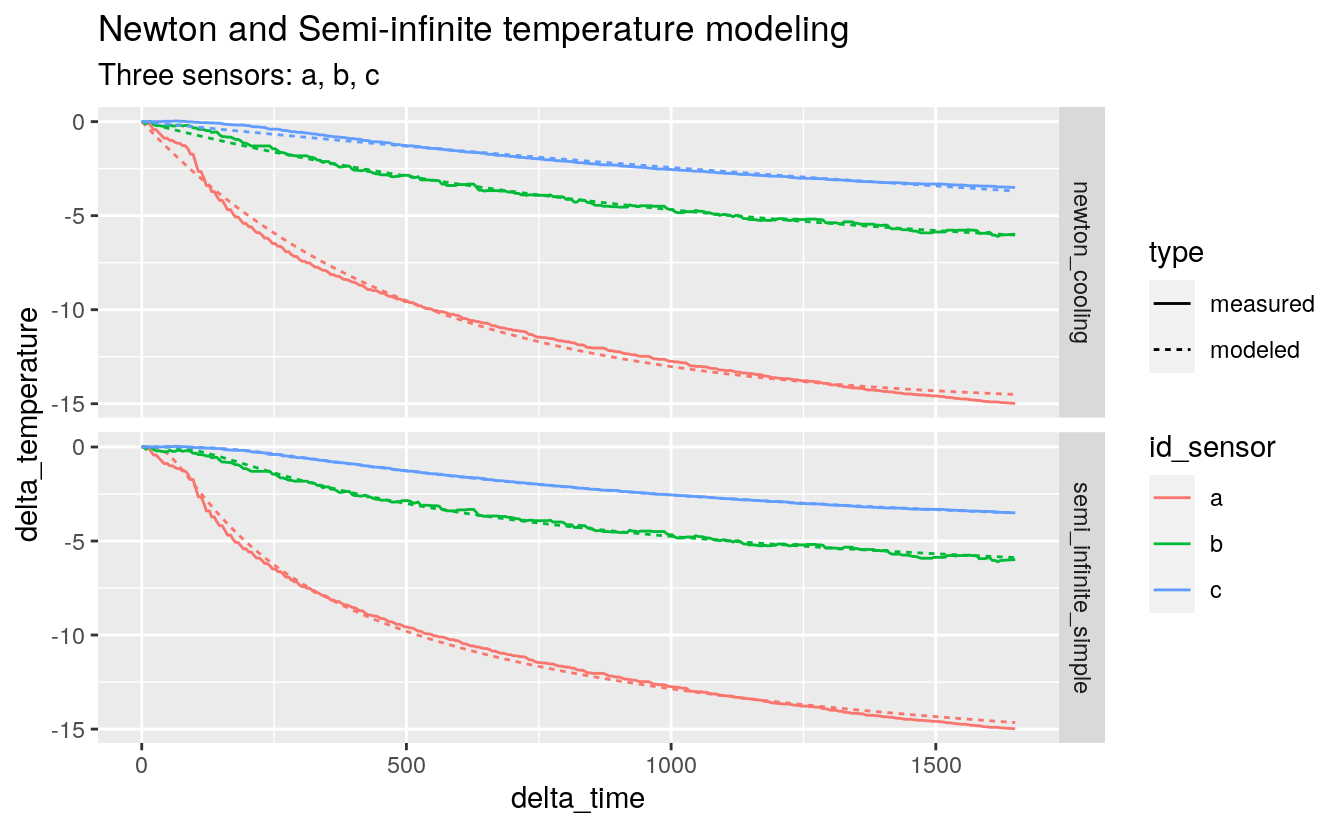
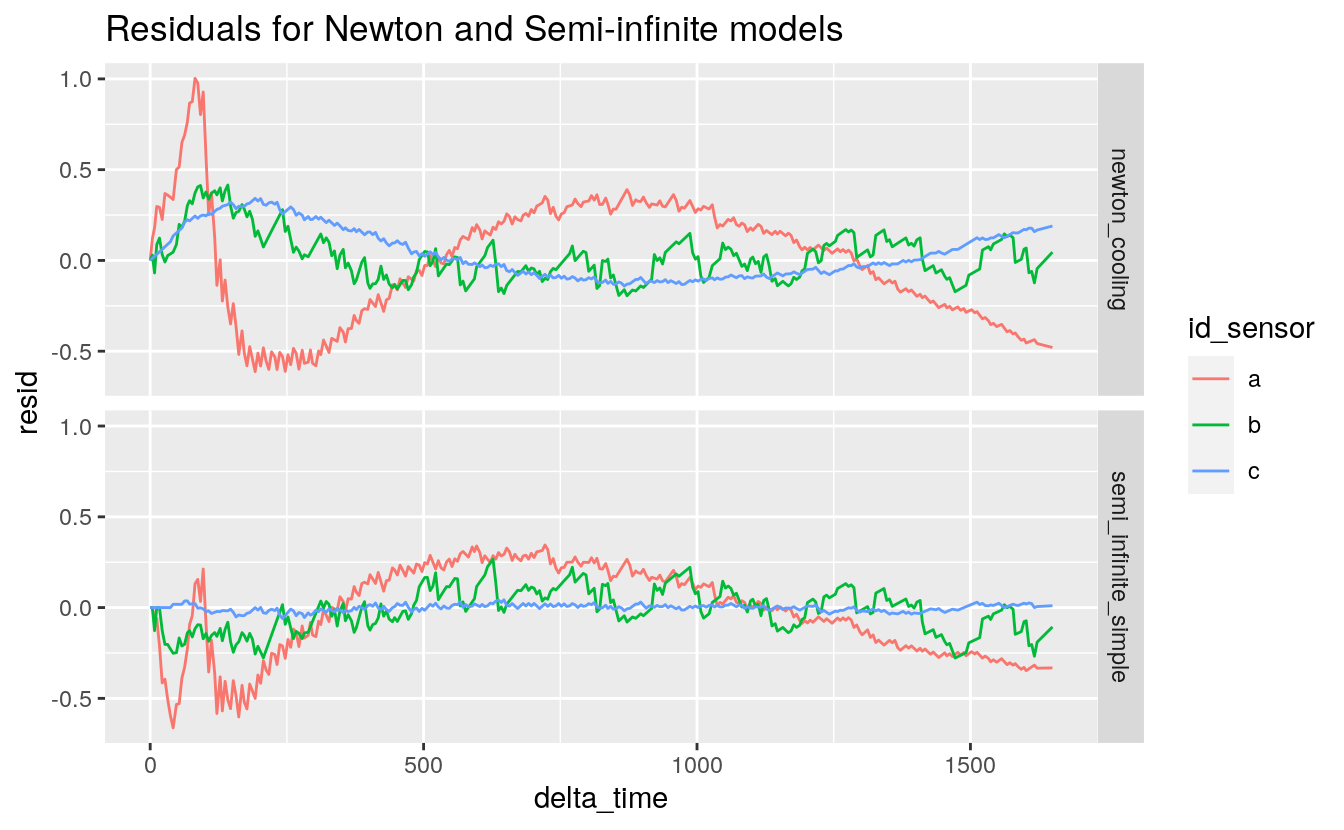
38.6.10 Let’s get the residuals
resid <-
model_nested_new %>%
mutate(resid = map(model, resid)) %>%
unnest(data, resid) %>%
print()
#> Warning: unnest() has a new interface. See ?unnest for details.
#> Try `df %>% unnest(c(data, resid))`, with `mutate()` if needed
#> # A tibble: 1,962 x 6
#> # Groups: id_sensor [3]
#> id_model id_sensor delta_time delta_temperature model resid
#> <chr> <chr> <dbl> <dbl> <list> <dbl>
#> 1 newton_cooling a 0 0 <nls> 0
#> 2 newton_cooling a 4 0 <nls> 0.120
#> 3 newton_cooling a 8 -0.06 <nls> 0.179
#> 4 newton_cooling a 12 -0.06 <nls> 0.297
#> 5 newton_cooling a 17 -0.211 <nls> 0.292
#> 6 newton_cooling a 22 -0.423 <nls> 0.225
#> # … with 1,956 more rows38.7 Using broom package to look at model-statistics
We will use a previous defined dataframe with the model and data:
model_nested_new %>%
print()
#> # A tibble: 6 x 4
#> # Groups: id_sensor [3]
#> id_model id_sensor data model
#> <chr> <chr> <list> <list>
#> 1 newton_cooling a <tibble [327 × 2]> <nls>
#> 2 newton_cooling b <tibble [327 × 2]> <nls>
#> 3 newton_cooling c <tibble [327 × 2]> <nls>
#> 4 semi_infinite_simple a <tibble [327 × 2]> <nls>
#> 5 semi_infinite_simple b <tibble [327 × 2]> <nls>
#> 6 semi_infinite_simple c <tibble [327 × 2]> <nls>The tidy() function extracts statistics from a model.
# apply over model_nested_new but only three variables
model_parameters <-
model_nested_new %>%
select(id_model, id_sensor, model) %>%
mutate(tidy = map(model, tidy)) %>%
select(-model) %>%
unnest() %>%
print()
#> Warning: `cols` is now required.
#> Please use `cols = c(tidy)`
#> # A tibble: 12 x 7
#> # Groups: id_sensor [3]
#> id_model id_sensor term estimate std.error statistic p.value
#> <chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl>
#> 1 newton_cooli… a delta_temperat… -15.1 0.0526 -286. 0.
#> 2 newton_cooli… a tau_0 500. 4.84 103. 1.07e-250
#> 3 newton_cooli… b delta_temperat… -7.59 0.0676 -112. 6.38e-262
#> 4 newton_cooli… b tau_0 1041. 16.2 64.2 9.05e-187
#> 5 newton_cooli… c delta_temperat… -9.87 0.704 -14.0 3.16e- 35
#> 6 newton_cooli… c tau_0 3525. 299. 11.8 5.61e- 27
#> # … with 6 more rows38.7.1 Get a sense of the coefficients
model_summary <-
model_parameters %>%
select(id_model, id_sensor, term, estimate) %>%
spread(key = "term", value = "estimate") %>%
print()
#> # A tibble: 6 x 4
#> # Groups: id_sensor [3]
#> id_model id_sensor delta_temperature_0 tau_0
#> <chr> <chr> <dbl> <dbl>
#> 1 newton_cooling a -15.1 500.
#> 2 newton_cooling b -7.59 1041.
#> 3 newton_cooling c -9.87 3525.
#> 4 semi_infinite_simple a -21.5 139.
#> 5 semi_infinite_simple b -10.6 287.
#> 6 semi_infinite_simple c -8.04 500.38.7.2 Summary
this is just a smalll part of purrr
-
there seem to be parallels between
tidyr::nest()/purrr::map()anddplyr::group_by()/dplyr::do()- to my mind, the purrr framework is more understandable
- update tweet from Hadley
References from Hadley: